Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1048-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1048-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 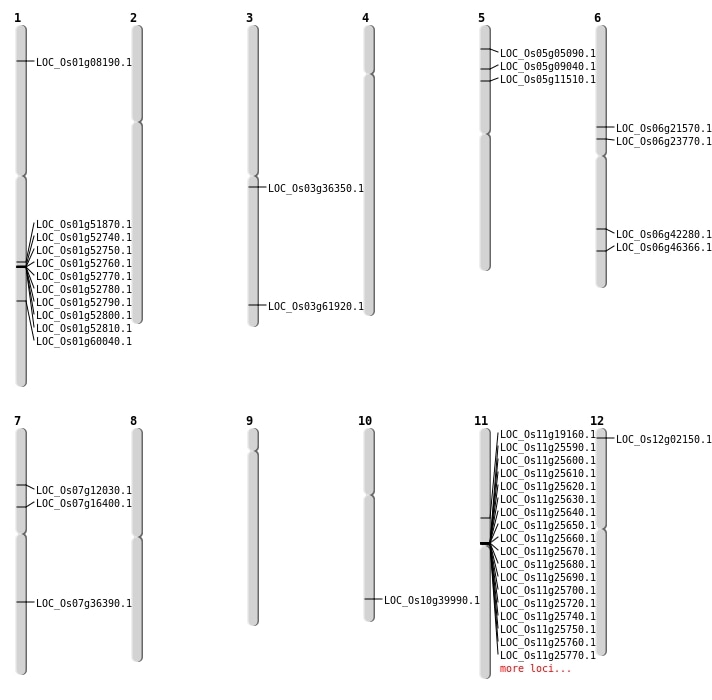
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10356436 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 20863020 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 22631028 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 23808945 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25399183 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 25399184 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 29820835 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g51870 |
| SBS | Chr1 | 3976504 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g08190 |
| SBS | Chr11 | 16627096 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 26593779 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44014 |
| SBS | Chr12 | 10802794 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 2789164 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 22243344 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 22899544 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 30361514 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 5527166 | T-A | HOMO | INTRON | |
| SBS | Chr3 | 17820005 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 20158363 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g36350 |
| SBS | Chr3 | 24447501 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 33055081 | A-T | HET | INTRON | |
| SBS | Chr3 | 35100928 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g61920 |
| SBS | Chr4 | 12415569 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 26393897 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 33801963 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 2473184 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g05090 |
| SBS | Chr5 | 27566594 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 5016518 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09040 |
| SBS | Chr6 | 10003129 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 12428579 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 14927524 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 15066808 | A-G | HET | INTRON | |
| SBS | Chr6 | 15745350 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 25148430 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 25390116 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42280 |
| SBS | Chr7 | 10404960 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 1065828 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 27294492 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 3326238 | C-G | HET | INTRON | |
| SBS | Chr7 | 7898368 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 9598076 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16400 |
| SBS | Chr8 | 12712852 | T-C | HET | INTRON | |
| SBS | Chr8 | 20923220 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 7644392 | T-C | HOMO | INTRON | |
| SBS | Chr9 | 12081052 | T-C | HOMO | INTRON | |
| SBS | Chr9 | 7877999 | G-A | HOMO | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 484056 | 484077 | 21 | |
| Deletion | Chr12 | 658166 | 658170 | 4 | LOC_Os12g02150 |
| Deletion | Chr3 | 6307689 | 6307690 | 1 | |
| Deletion | Chr7 | 6720252 | 6720258 | 6 | LOC_Os07g12030 |
| Deletion | Chr6 | 7128848 | 7128861 | 13 | |
| Deletion | Chr2 | 7931565 | 7931570 | 5 | |
| Deletion | Chr5 | 8549394 | 8549395 | 1 | |
| Deletion | Chr2 | 10863732 | 10863733 | 1 | |
| Deletion | Chr5 | 11980262 | 11980270 | 8 | |
| Deletion | Chr6 | 12441788 | 12444151 | 2363 | LOC_Os06g21570 |
| Deletion | Chr6 | 13885991 | 13886001 | 10 | LOC_Os06g23770 |
| Deletion | Chr6 | 13946684 | 13946719 | 35 | |
| Deletion | Chr11 | 14595001 | 15235000 | 640000 | 92 |
| Deletion | Chr11 | 17535697 | 17535710 | 13 | |
| Deletion | Chr6 | 20974975 | 20974976 | 1 | |
| Deletion | Chr10 | 21424402 | 21424406 | 4 | LOC_Os10g39990 |
| Deletion | Chr7 | 21755066 | 21755072 | 6 | LOC_Os07g36390 |
| Deletion | Chr6 | 22277845 | 22277852 | 7 | |
| Deletion | Chr11 | 25421562 | 25421594 | 32 | |
| Deletion | Chr12 | 27164289 | 27164294 | 5 | |
| Deletion | Chr4 | 30070454 | 30070463 | 9 | |
| Deletion | Chr1 | 30335001 | 30382000 | 47000 | 8 |
| Deletion | Chr1 | 31275039 | 31275042 | 3 | |
| Deletion | Chr1 | 31635402 | 31635403 | 1 | |
| Deletion | Chr4 | 32833417 | 32833418 | 1 | |
| Deletion | Chr3 | 33117326 | 33117357 | 31 | |
| Deletion | Chr4 | 33570561 | 33570562 | 1 | |
| Deletion | Chr1 | 34707330 | 34707342 | 12 | LOC_Os01g60040 |
| Deletion | Chr4 | 34739460 | 34739462 | 2 | |
| Deletion | Chr1 | 42473455 | 42473459 | 4 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 17838567 | 17838570 | 4 | |
| Insertion | Chr11 | 17165681 | 17165681 | 1 | |
| Insertion | Chr4 | 20326665 | 20326665 | 1 | |
| Insertion | Chr6 | 28133805 | 28133806 | 2 | LOC_Os06g46366 |
| Insertion | Chr8 | 19998614 | 19998614 | 1 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6516207 | 6585935 | LOC_Os05g11510 |
| Inversion | Chr5 | 6524232 | 6585941 | |
| Inversion | Chr11 | 7903490 | 9827468 | |
| Inversion | Chr11 | 8875221 | 10935775 | LOC_Os11g19160 |
| Inversion | Chr11 | 9823939 | 9828521 | |
| Inversion | Chr11 | 14580179 | 15341443 | |
| Inversion | Chr11 | 15235102 | 15341477 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 9574205 | Chr1 | 6539783 | |
| Translocation | Chr6 | 12441794 | Chr3 | 4348338 | LOC_Os06g21570 |
| Translocation | Chr6 | 12444143 | Chr3 | 4348298 | |
| Translocation | Chr11 | 19839718 | Chr4 | 2841993 | LOC_Os11g33942 |
