Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1060-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1060-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 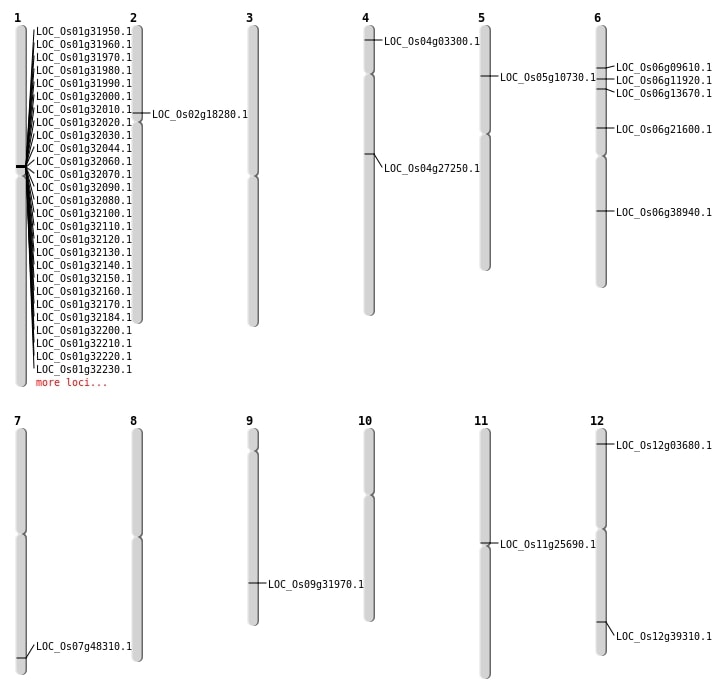
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19546562 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 39624507 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 14090155 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11107428 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 15214155 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 15290807 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 25515799 | T-C | HET | INTRON | |
| SBS | Chr2 | 10600531 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18280 |
| SBS | Chr2 | 31702300 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 33267463 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 951100 | T-A | HET | INTRON | |
| SBS | Chr3 | 18781559 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 21883221 | T-G | HOMO | INTRON | |
| SBS | Chr3 | 33127137 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 7550098 | T-C | HET | INTRON | |
| SBS | Chr3 | 7550099 | T-C | HET | INTRON | |
| SBS | Chr4 | 27654242 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 6435231 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 15517336 | G-A | HOMO | INTRON | |
| SBS | Chr5 | 15658304 | A-G | HET | INTRON | |
| SBS | Chr5 | 29891791 | T-G | HOMO | INTRON | |
| SBS | Chr5 | 549263 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 6555568 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 8320712 | C-T | HET | INTRON | |
| SBS | Chr6 | 183937 | C-T | HET | INTRON | |
| SBS | Chr6 | 20452463 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 27490856 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 4890513 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g09610 |
| SBS | Chr6 | 4890530 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 13647820 | T-C | HET | INTRON | |
| SBS | Chr8 | 11636722 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 2157407 | G-A | HET | INTRON | |
| SBS | Chr8 | 8455188 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr9 | 12403577 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12404585 | A-T | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1812003 | 1812005 | 2 | |
| Deletion | Chr5 | 3701946 | 3701981 | 35 | |
| Deletion | Chr5 | 5910215 | 5910216 | 1 | LOC_Os05g10730 |
| Deletion | Chr6 | 6349146 | 6349147 | 1 | LOC_Os06g11920 |
| Deletion | Chr9 | 6376505 | 6376514 | 9 | |
| Deletion | Chr4 | 6578777 | 6578786 | 9 | |
| Deletion | Chr5 | 7091674 | 7091686 | 12 | |
| Deletion | Chr6 | 7587193 | 7587530 | 337 | |
| Deletion | Chr12 | 11819267 | 11819280 | 13 | |
| Deletion | Chr6 | 12457313 | 12457320 | 7 | LOC_Os06g21600 |
| Deletion | Chr11 | 14653116 | 14653134 | 18 | LOC_Os11g25690 |
| Deletion | Chr9 | 16615113 | 16615125 | 12 | |
| Deletion | Chr1 | 17494001 | 17852000 | 358000 | 54 |
| Deletion | Chr8 | 18539610 | 18539615 | 5 | |
| Deletion | Chr7 | 19185490 | 19185504 | 14 | |
| Deletion | Chr11 | 21127686 | 21127693 | 7 | |
| Deletion | Chr7 | 21929272 | 21929274 | 2 | |
| Deletion | Chr6 | 23101963 | 23101965 | 2 | LOC_Os06g38940 |
| Deletion | Chr12 | 24186535 | 24186551 | 16 | LOC_Os12g39310 |
| Deletion | Chr6 | 26971128 | 26971139 | 11 | |
| Deletion | Chr6 | 27032175 | 27032201 | 26 | |
| Deletion | Chr7 | 28864404 | 28864405 | 1 | LOC_Os07g48310 |
| Deletion | Chr1 | 38441013 | 38441019 | 6 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 16099590 | 16099605 | 16 |
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1482576 | Chr2 | 9635648 | LOC_Os12g03680 |
| Translocation | Chr8 | 6896137 | Chr4 | 1405353 | LOC_Os04g03300 |
| Translocation | Chr5 | 8774636 | Chr4 | 16102013 | LOC_Os04g27250 |
| Translocation | Chr9 | 19075659 | Chr6 | 7587521 | 2 |
| Translocation | Chr9 | 19076131 | Chr6 | 7587216 | 2 |
