Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1075-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1075-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 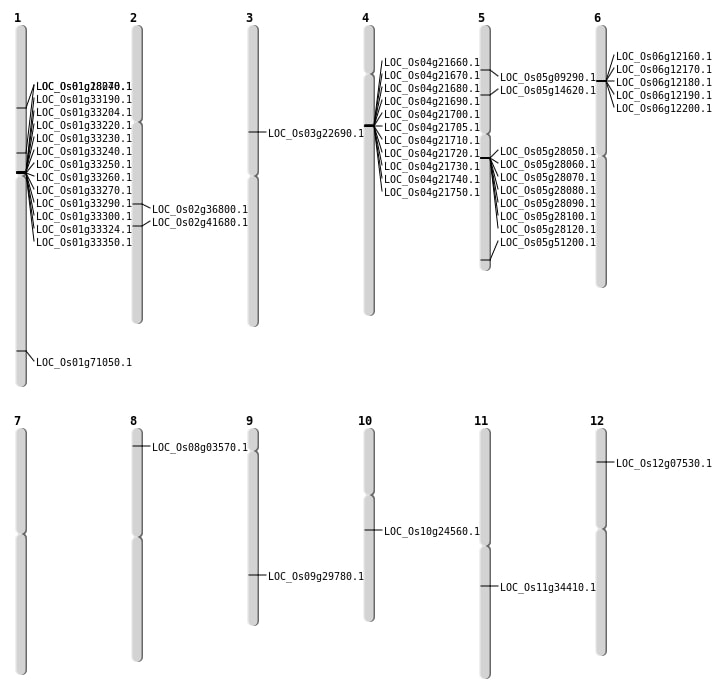
|
| Variant Information |
|---|
Single base substitutions: 52
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10093467 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g18040 |
| SBS | Chr1 | 11559019 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 15838395 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g28270 |
| SBS | Chr1 | 17952043 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 26561629 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 32893070 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 41117889 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g71050 |
| SBS | Chr10 | 12604490 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24560 |
| SBS | Chr10 | 21486934 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 4015204 | T-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 10682245 | A-T | HET | INTRON | |
| SBS | Chr11 | 2667818 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 332935 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 5869009 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 11610628 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 20537786 | G-A | HET | INTRON | |
| SBS | Chr2 | 20895932 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 22196076 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g36800 |
| SBS | Chr2 | 24501257 | C-T | HET | INTRON | |
| SBS | Chr2 | 34022808 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 34586845 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 13108725 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g22690 |
| SBS | Chr3 | 20791469 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28083227 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 30106230 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 5250028 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 24906049 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 11752413 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 17768712 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 19963032 | T-G | HET | UTR_5_PRIME | |
| SBS | Chr5 | 5190180 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09290 |
| SBS | Chr5 | 8328982 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14620 |
| SBS | Chr5 | 9040212 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 10610156 | T-A | HET | INTRON | |
| SBS | Chr6 | 17934547 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 21846001 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 26600582 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 30894083 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 30969048 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 994243 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 235830 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 26445279 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8117095 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 8680791 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 1318622 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 16451361 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 8303236 | C-A | HET | INTRON | |
| SBS | Chr9 | 10010598 | A-G | HET | INTRON | |
| SBS | Chr9 | 11688937 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 15810953 | G-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 659188 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 854260 | T-C | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1670622 | 1670631 | 9 | LOC_Os08g03570 |
| Deletion | Chr10 | 5028307 | 5028314 | 7 | |
| Deletion | Chr2 | 6000329 | 6000334 | 5 | |
| Deletion | Chr10 | 6192772 | 6192822 | 50 | |
| Deletion | Chr11 | 6289689 | 6289712 | 23 | |
| Deletion | Chr6 | 6513001 | 6556000 | 43000 | 5 |
| Deletion | Chr12 | 9968414 | 9968415 | 1 | |
| Deletion | Chr8 | 11418342 | 11418343 | 1 | |
| Deletion | Chr4 | 12255954 | 12311893 | 55939 | 11 |
| Deletion | Chr5 | 16392001 | 16446000 | 54000 | 7 |
| Deletion | Chr6 | 16466740 | 16467090 | 350 | |
| Deletion | Chr11 | 17080461 | 17080462 | 1 | |
| Deletion | Chr1 | 17386223 | 17386225 | 2 | |
| Deletion | Chr9 | 18098936 | 18098946 | 10 | LOC_Os09g29780 |
| Deletion | Chr1 | 18273001 | 18370000 | 97000 | 12 |
| Deletion | Chr12 | 18307813 | 18307832 | 19 | |
| Deletion | Chr11 | 20162928 | 20162932 | 4 | LOC_Os11g34410 |
| Deletion | Chr10 | 22047190 | 22047195 | 5 | |
| Deletion | Chr2 | 25007731 | 25007737 | 6 | LOC_Os02g41680 |
| Deletion | Chr5 | 25828951 | 25828953 | 2 | |
| Deletion | Chr4 | 30672721 | 30672724 | 3 | |
| Deletion | Chr1 | 31977079 | 31977085 | 6 | |
| Deletion | Chr1 | 32444859 | 32444861 | 2 | |
| Deletion | Chr1 | 34312130 | 34312162 | 32 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 3646191 | 3747092 | LOC_Os12g07530 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 16466770 | Chr5 | 29370720 | LOC_Os05g51200 |
| Translocation | Chr6 | 16467079 | Chr5 | 29505342 |
