Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1076-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1076-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 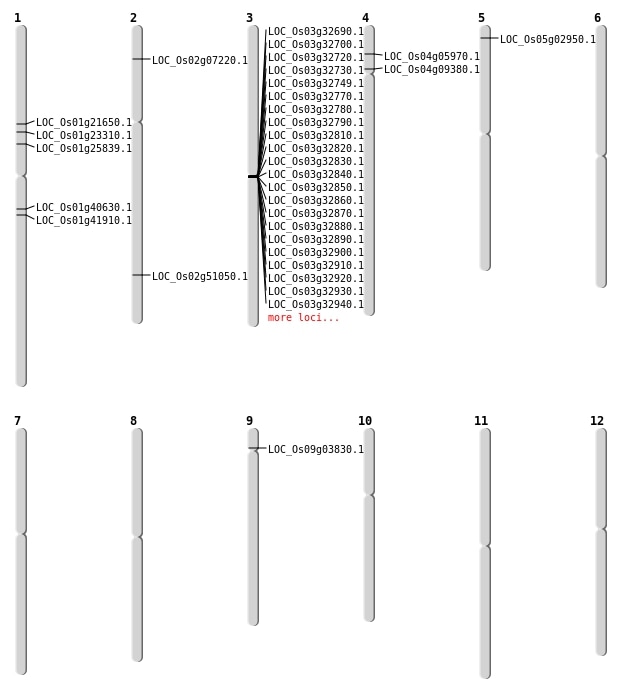
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10381556 | C-T | HET | INTRON | |
| SBS | Chr1 | 12130833 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g21650 |
| SBS | Chr1 | 23771061 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41910 |
| SBS | Chr1 | 25369840 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 40502152 | C-T | HET | INTRON | |
| SBS | Chr10 | 10638516 | A-G | HET | INTRON | |
| SBS | Chr10 | 16370041 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 20981599 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 7446959 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 31226574 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g51050 |
| SBS | Chr3 | 17146514 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 25225033 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 27067163 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 30904379 | G-A | HET | INTRON | |
| SBS | Chr3 | 30904725 | A-T | HET | INTRON | |
| SBS | Chr3 | 3120045 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 17086786 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 20735769 | C-G | HOMO | INTRON | |
| SBS | Chr4 | 3090101 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05970 |
| SBS | Chr5 | 1094161 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g02950 |
| SBS | Chr5 | 13933874 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 18410515 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 26215187 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 3801182 | A-G | HET | INTRON | |
| SBS | Chr6 | 13986799 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 1681340 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 22418075 | G-T | HOMO | INTRON | |
| SBS | Chr6 | 2474429 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 13068842 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 1941112 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g03830 |
| SBS | Chr9 | 20871298 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 21074352 | C-T | HET | SYNONYMOUS_CODING |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 3711454 | 3711457 | 3 | LOC_Os02g07220 |
| Deletion | Chr4 | 4273185 | 4273188 | 3 | |
| Deletion | Chr7 | 5806315 | 5806317 | 2 | |
| Deletion | Chr10 | 7816362 | 7816373 | 11 | |
| Deletion | Chr2 | 7925107 | 7925137 | 30 | |
| Deletion | Chr12 | 10471781 | 10471782 | 1 | |
| Deletion | Chr7 | 13217634 | 13217649 | 15 | |
| Deletion | Chr11 | 13613698 | 13613700 | 2 | |
| Deletion | Chr1 | 14635774 | 14656786 | 21012 | LOC_Os01g25839 |
| Deletion | Chr1 | 14912452 | 14912453 | 1 | |
| Deletion | Chr7 | 15064389 | 15064390 | 1 | |
| Deletion | Chr10 | 18171501 | 18171520 | 19 | |
| Deletion | Chr3 | 18715001 | 19068000 | 353000 | 61 |
| Deletion | Chr5 | 18745726 | 18745734 | 8 | |
| Deletion | Chr7 | 19576627 | 19576628 | 1 | |
| Deletion | Chr3 | 22663565 | 22663566 | 1 | LOC_Os03g40725 |
| Deletion | Chr3 | 23568476 | 23568482 | 6 | LOC_Os03g42370 |
| Deletion | Chr11 | 23974342 | 23974360 | 18 | |
| Deletion | Chr6 | 25156304 | 25156308 | 4 |
Insertions: 9
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 1697763 | Chr1 | 22958758 | LOC_Os01g40630 |
| Translocation | Chr4 | 11225765 | Chr1 | 13093223 | LOC_Os01g23310 |
| Translocation | Chr7 | 17250533 | Chr4 | 5000289 | LOC_Os04g09380 |
