Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1080-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1080-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 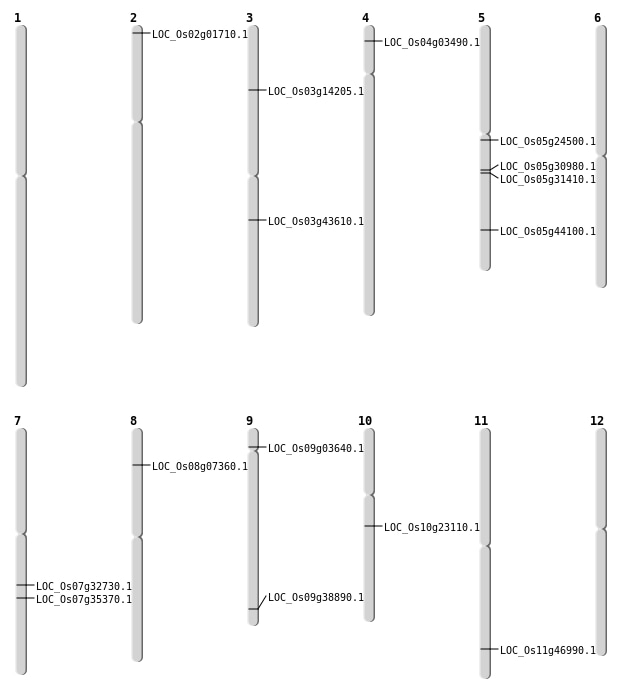
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12194057 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 16962520 | T-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr1 | 22904478 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 5587510 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12057585 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g23110 |
| SBS | Chr11 | 13169703 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 21328438 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 2066555 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3824476 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20541601 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 23366033 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 23658613 | A-T | HET | INTRON | |
| SBS | Chr2 | 25215812 | G-A | HET | INTRON | |
| SBS | Chr2 | 6142240 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 8906321 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 1009729 | T-C | HET | INTRON | |
| SBS | Chr3 | 1009749 | A-T | HET | INTRON | |
| SBS | Chr3 | 10261920 | T-A | HET | INTRON | |
| SBS | Chr3 | 21382866 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 21568470 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 24374368 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g43610 |
| SBS | Chr3 | 28923535 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 10061202 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 1511023 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03490 |
| SBS | Chr4 | 28766209 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 33069023 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 8757671 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 11352027 | G-A | HET | INTRON | |
| SBS | Chr5 | 25643240 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44100 |
| SBS | Chr6 | 14163617 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17385584 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 27451654 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 27451655 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 13776145 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 19603789 | C-G | HOMO | INTRON | |
| SBS | Chr7 | 25951068 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 26879848 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 5223040 | C-A | HOMO | INTRON | |
| SBS | Chr8 | 12103181 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 4123060 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g07360 |
| SBS | Chr9 | 1826990 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g03640 |
| SBS | Chr9 | 22331835 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38890 |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 394364 | 394368 | 4 | LOC_Os02g01710 |
| Deletion | Chr5 | 638062 | 638133 | 71 | |
| Deletion | Chr5 | 2131594 | 2131596 | 2 | |
| Deletion | Chr3 | 6789116 | 6789125 | 9 | |
| Deletion | Chr12 | 10828612 | 10828615 | 3 | |
| Deletion | Chr11 | 11053115 | 11053116 | 1 | |
| Deletion | Chr4 | 11426929 | 11426941 | 12 | |
| Deletion | Chr4 | 11505525 | 11505538 | 13 | |
| Deletion | Chr3 | 12810397 | 12810404 | 7 | |
| Deletion | Chr7 | 19603742 | 19603744 | 2 | |
| Deletion | Chr12 | 22492494 | 22492521 | 27 | |
| Deletion | Chr1 | 23223832 | 23223833 | 1 | |
| Deletion | Chr6 | 26584308 | 26584309 | 1 | |
| Deletion | Chr11 | 28242167 | 28242170 | 3 | LOC_Os11g46990 |
| Deletion | Chr5 | 29077691 | 29077701 | 10 | |
| Deletion | Chr3 | 35760288 | 35760327 | 39 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12682472 | 12682472 | 1 | |
| Insertion | Chr10 | 10335433 | 10335433 | 1 | |
| Insertion | Chr3 | 7707175 | 7707183 | 9 | LOC_Os03g14205 |
| Insertion | Chr7 | 27877529 | 27877529 | 1 | |
| Insertion | Chr8 | 22265044 | 22265044 | 1 | |
| Insertion | Chr9 | 10760409 | 10760414 | 6 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 8755148 | 8852729 | |
| Inversion | Chr5 | 14034814 | 14141044 | LOC_Os05g24500 |
| Inversion | Chr5 | 18000375 | 18265182 | 2 |
| Inversion | Chr5 | 18000401 | 18265692 | LOC_Os05g30980 |
| Inversion | Chr5 | 18265387 | 18752043 | LOC_Os05g31410 |
| Inversion | Chr5 | 18265684 | 18752043 | |
| Inversion | Chr7 | 19544575 | 21155691 | 2 |
| Inversion | Chr7 | 19551818 | 21155704 | LOC_Os07g35370 |
| Inversion | Chr5 | 26352463 | 26359062 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8755134 | Chr5 | 25363240 | |
| Translocation | Chr12 | 8840820 | Chr7 | 19482301 | |
| Translocation | Chr9 | 8852715 | Chr5 | 25364224 |
