Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1085-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1085-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 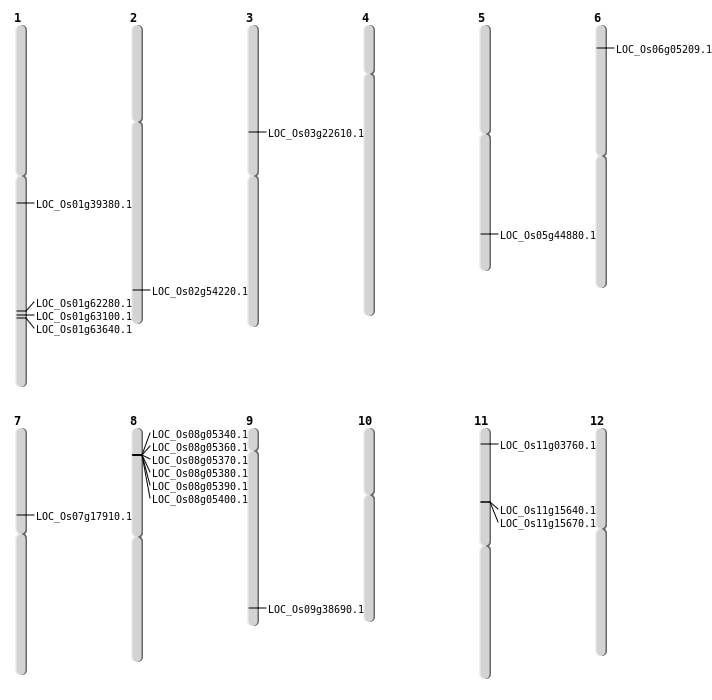
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16746424 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 22164879 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39380 |
| SBS | Chr1 | 36569064 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63100 |
| SBS | Chr1 | 36910778 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g63640 |
| SBS | Chr11 | 17477102 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1865810 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 8927668 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 9115770 | G-A | HET | INTRON | |
| SBS | Chr12 | 26966844 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 18911614 | C-T | HET | INTRON | |
| SBS | Chr2 | 33241532 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g54220 |
| SBS | Chr2 | 8185325 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10539211 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 17103186 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 24512691 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31045822 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 8077393 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 8077394 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 13103088 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 32291668 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 35403111 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 10702693 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 29582036 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 8128413 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17731344 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 23833534 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 29569577 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 10594479 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17910 |
| SBS | Chr7 | 21276557 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 6950256 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 8047538 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12272649 | C-A | HOMO | UTR_5_PRIME | |
| SBS | Chr8 | 13219128 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 15589488 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 20094347 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 5425612 | T-A | HET | INTRON | |
| SBS | Chr9 | 18752368 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr9 | 22241545 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38690 |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 622350 | 622364 | 14 | |
| Deletion | Chr6 | 2337555 | 2337568 | 13 | LOC_Os06g05209 |
| Deletion | Chr8 | 2813001 | 2850000 | 37000 | 6 |
| Deletion | Chr12 | 3766896 | 3766909 | 13 | |
| Deletion | Chr9 | 4344011 | 4344012 | 1 | |
| Deletion | Chr8 | 6502753 | 6502768 | 15 | |
| Deletion | Chr1 | 7103768 | 7103778 | 10 | |
| Deletion | Chr12 | 7256285 | 7256289 | 4 | |
| Deletion | Chr7 | 8021370 | 8021400 | 30 | |
| Deletion | Chr6 | 8168748 | 8168752 | 4 | |
| Deletion | Chr5 | 8558035 | 8558041 | 6 | |
| Deletion | Chr9 | 10781390 | 10781392 | 2 | |
| Deletion | Chr3 | 13033534 | 13033541 | 7 | LOC_Os03g22610 |
| Deletion | Chr9 | 13731036 | 13731043 | 7 | |
| Deletion | Chr11 | 14219313 | 14219314 | 1 | |
| Deletion | Chr1 | 14369435 | 14369437 | 2 | |
| Deletion | Chr5 | 15083582 | 15083587 | 5 | |
| Deletion | Chr2 | 19922142 | 19922151 | 9 | |
| Deletion | Chr8 | 20764814 | 20764816 | 2 | |
| Deletion | Chr12 | 24292606 | 24292607 | 1 | |
| Deletion | Chr5 | 26083815 | 26083826 | 11 | LOC_Os05g44880 |
| Deletion | Chr3 | 26622283 | 26622297 | 14 | |
| Deletion | Chr4 | 27384558 | 27384582 | 24 | |
| Deletion | Chr6 | 30272957 | 30272958 | 1 | |
| Deletion | Chr1 | 40035227 | 40035231 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 36038941 | 36038970 | 30 | LOC_Os01g62280 |
| Insertion | Chr12 | 25804867 | 25804870 | 4 | |
| Insertion | Chr4 | 16152276 | 16152276 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 1477004 | 8885353 | 2 |
| Inversion | Chr11 | 1477317 | 8873327 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 21189217 | Chr4 | 13727663 |
