Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1190-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1190-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 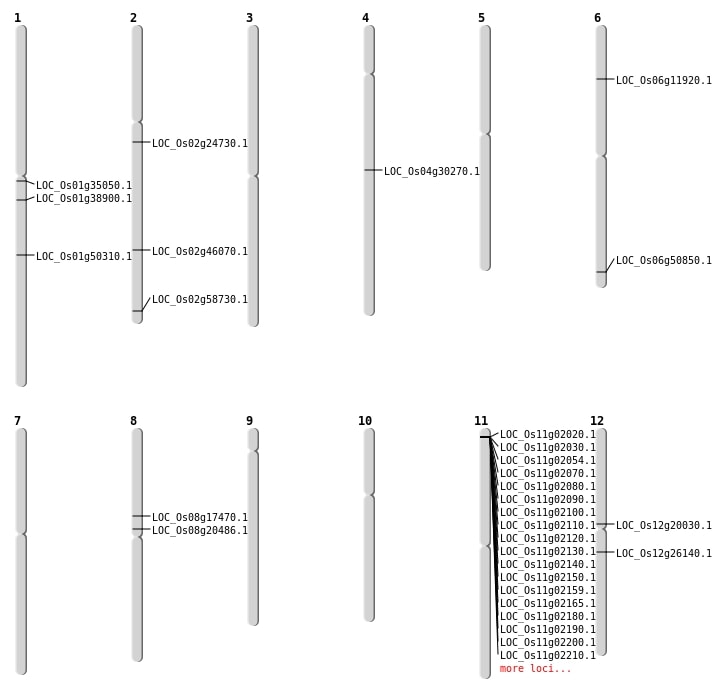
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19095241 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 21709566 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 21709567 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7469998 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 17752166 | G-T | HET | INTRON | |
| SBS | Chr10 | 3574721 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 25437102 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 5941325 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 11650749 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g20030 |
| SBS | Chr12 | 15223532 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26140 |
| SBS | Chr2 | 14345632 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24730 |
| SBS | Chr2 | 19745141 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 22255594 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 28074908 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g46070 |
| SBS | Chr2 | 659588 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 36121188 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 18071945 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g30270 |
| SBS | Chr4 | 31844194 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 33445058 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 7301078 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 27022138 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 2900965 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 10650284 | G-A | HET | INTRON | |
| SBS | Chr6 | 30770403 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g50850 |
| SBS | Chr6 | 6248225 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 6350872 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g11920 |
| SBS | Chr7 | 29572152 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 10194980 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 12294015 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g20486 |
| SBS | Chr8 | 22337643 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 27624684 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 27900836 | C-T | HET | INTRON | |
| SBS | Chr9 | 16038415 | G-A | HOMO | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 540001 | 1206000 | 666000 | 124 |
| Deletion | Chr3 | 2629234 | 2629239 | 5 | |
| Deletion | Chr2 | 5557544 | 5557546 | 2 | |
| Deletion | Chr6 | 5687541 | 5687548 | 7 | |
| Deletion | Chr11 | 6460383 | 6460384 | 1 | |
| Deletion | Chr6 | 6532043 | 6532048 | 5 | |
| Deletion | Chr8 | 10699285 | 10699292 | 7 | LOC_Os08g17470 |
| Deletion | Chr2 | 12857534 | 12857539 | 5 | |
| Deletion | Chr12 | 13233323 | 13233340 | 17 | |
| Deletion | Chr2 | 14811820 | 14811827 | 7 | |
| Deletion | Chr10 | 18151795 | 18151800 | 5 | |
| Deletion | Chr8 | 19995140 | 19995141 | 1 | |
| Deletion | Chr5 | 21123335 | 21123343 | 8 | |
| Deletion | Chr7 | 22375945 | 22375948 | 3 | |
| Deletion | Chr5 | 24010764 | 24010781 | 17 | |
| Deletion | Chr7 | 26496646 | 26496662 | 16 | |
| Deletion | Chr5 | 26825473 | 26825500 | 27 | |
| Deletion | Chr8 | 28206687 | 28206708 | 21 | |
| Deletion | Chr1 | 28881601 | 28881603 | 2 | LOC_Os01g50310 |
| Deletion | Chr1 | 29364110 | 29364111 | 1 | |
| Deletion | Chr2 | 35885397 | 35885399 | 2 | LOC_Os02g58730 |
| Deletion | Chr1 | 41776204 | 41776205 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 13912052 | 13912054 | 3 | |
| Insertion | Chr5 | 6514541 | 6514541 | 1 | |
| Insertion | Chr5 | 8610327 | 8610327 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 530948 | 539368 | |
| Inversion | Chr11 | 530960 | 1205773 | LOC_Os11g03270 |
| Inversion | Chr1 | 19401515 | 21843892 | 2 |
No Translocation
