Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1206-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1206-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 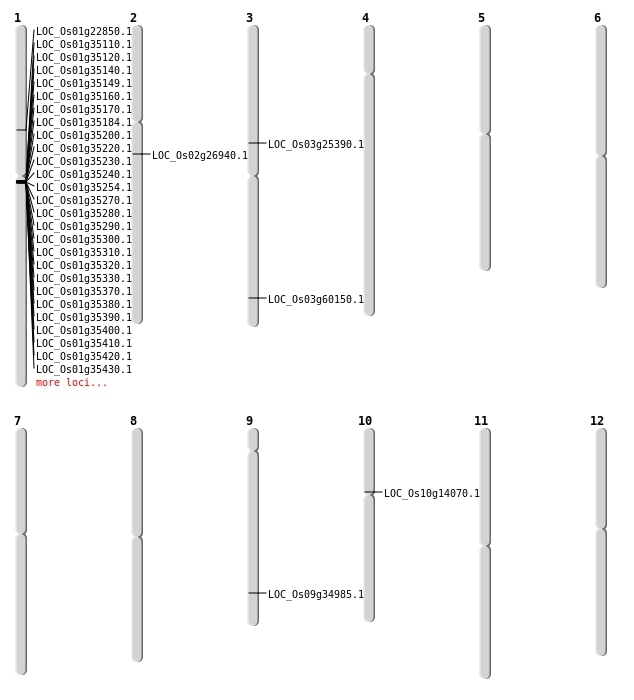
|
| Variant Information |
|---|
Single base substitutions: 18
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16606668 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 21765505 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 38720334 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 43081306 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7656358 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g14070 |
| SBS | Chr2 | 17764226 | A-C | HET | INTRON | |
| SBS | Chr3 | 23416423 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 26720599 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 14682344 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2289633 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 30040177 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 11483208 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 18899268 | C-T | HET | INTRON | |
| SBS | Chr5 | 19113351 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 7316176 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 22034754 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 10009156 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 12392977 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2303851 | 2303860 | 9 | |
| Deletion | Chr7 | 2935845 | 2935910 | 65 | |
| Deletion | Chr7 | 4419396 | 4419398 | 2 | |
| Deletion | Chr5 | 6644509 | 6644512 | 3 | |
| Deletion | Chr2 | 15721186 | 15721187 | 1 | |
| Deletion | Chr2 | 15823980 | 15823985 | 5 | LOC_Os02g26940 |
| Deletion | Chr1 | 17484869 | 17484870 | 1 | |
| Deletion | Chr1 | 19437001 | 19483000 | 46000 | 8 |
| Deletion | Chr1 | 19494001 | 19556000 | 62000 | 11 |
| Deletion | Chr1 | 19569001 | 19649000 | 80000 | 14 |
| Deletion | Chr8 | 19995140 | 19995141 | 1 | |
| Deletion | Chr9 | 20379507 | 20379519 | 12 | LOC_Os09g34985 |
| Deletion | Chr1 | 21904398 | 21904400 | 2 | |
| Deletion | Chr4 | 22086859 | 22086877 | 18 | |
| Deletion | Chr2 | 25142195 | 25142200 | 5 | |
| Deletion | Chr2 | 25915485 | 25915490 | 5 | |
| Deletion | Chr1 | 26069491 | 26069493 | 2 | |
| Deletion | Chr5 | 26746029 | 26746030 | 1 | |
| Deletion | Chr7 | 29366494 | 29366496 | 2 | |
| Deletion | Chr1 | 31371917 | 31372109 | 192 | LOC_Os01g54550 |
| Deletion | Chr3 | 34202617 | 34202623 | 6 | LOC_Os03g60150 |
| Deletion | Chr1 | 35618295 | 35618302 | 7 | |
| Deletion | Chr1 | 36755001 | 36818000 | 63000 | 6 |
| Deletion | Chr1 | 41525016 | 41525017 | 1 |
Insertions: 8
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 36510285 | 36755453 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 14517220 | Chr1 | 12839256 | 2 |
| Translocation | Chr9 | 15196259 | Chr1 | 31372150 | LOC_Os01g54550 |
