Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1227-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1227-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 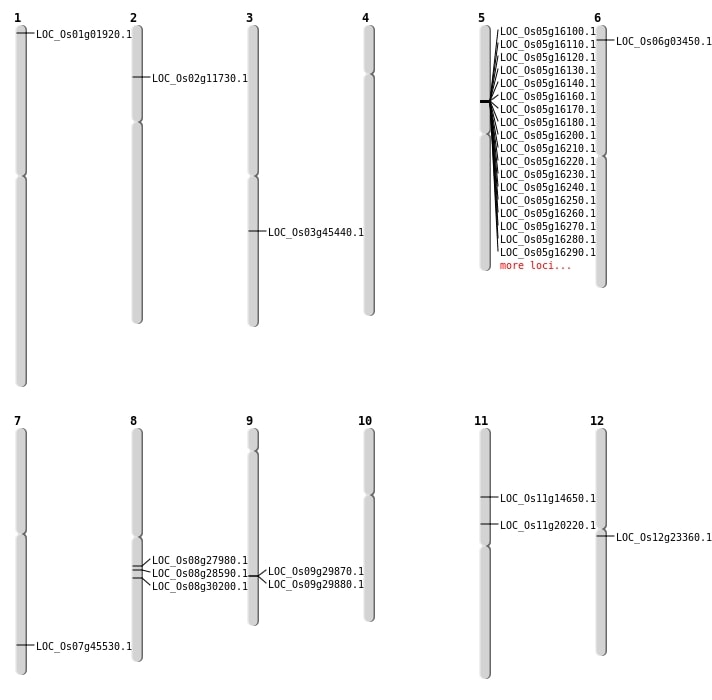
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15873160 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 27064524 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 12127743 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 13101687 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 20102496 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 22812170 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 6211157 | A-G | HET | INTRON | |
| SBS | Chr10 | 82899 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 90655 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 10293464 | G-A | HET | INTRON | |
| SBS | Chr11 | 11679980 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g20220 |
| SBS | Chr11 | 16034828 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8257313 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g14650 |
| SBS | Chr12 | 13220302 | G-A | HET | STOP_GAINED | LOC_Os12g23360 |
| SBS | Chr12 | 1350689 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 1350694 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 17353260 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 12773949 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 10363423 | A-C | HET | INTRON | |
| SBS | Chr4 | 15076645 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 21689251 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 3021364 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 12968172 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g22850 |
| SBS | Chr5 | 14965592 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 14965593 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26904105 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 8717667 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 12263672 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 1321357 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g03450 |
| SBS | Chr6 | 16038474 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29717888 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 7163284 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 23803932 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 28887714 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 29181488 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr8 | 16766788 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 17460443 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g28590 |
| SBS | Chr8 | 18562690 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30200 |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 186967 | 186973 | 6 | |
| Deletion | Chr1 | 338922 | 338926 | 4 | |
| Deletion | Chr1 | 506020 | 506025 | 5 | LOC_Os01g01920 |
| Deletion | Chr12 | 4425387 | 4425394 | 7 | |
| Deletion | Chr11 | 8672349 | 8672352 | 3 | |
| Deletion | Chr9 | 8836778 | 8836783 | 5 | |
| Deletion | Chr9 | 9061510 | 9061511 | 1 | |
| Deletion | Chr5 | 9096861 | 10344050 | 1247189 | 181 |
| Deletion | Chr8 | 11237246 | 11237247 | 1 | |
| Deletion | Chr6 | 12203800 | 12203801 | 1 | |
| Deletion | Chr11 | 12728550 | 12728558 | 8 | |
| Deletion | Chr5 | 13947001 | 14153000 | 206000 | 36 |
| Deletion | Chr6 | 16238779 | 16238786 | 7 | |
| Deletion | Chr5 | 16287221 | 16287222 | 1 | |
| Deletion | Chr8 | 17049350 | 17049357 | 7 | LOC_Os08g27980 |
| Deletion | Chr12 | 17338389 | 17338393 | 4 | |
| Deletion | Chr10 | 17576219 | 17576221 | 2 | |
| Deletion | Chr9 | 17779579 | 17779589 | 10 | |
| Deletion | Chr10 | 18069056 | 18069057 | 1 | |
| Deletion | Chr9 | 18160538 | 18173675 | 13137 | 2 |
| Deletion | Chr7 | 22856981 | 22856982 | 1 | |
| Deletion | Chr7 | 25476972 | 25476975 | 3 | |
| Deletion | Chr3 | 25648583 | 25649172 | 589 | LOC_Os03g45440 |
| Deletion | Chr5 | 25916561 | 25916570 | 9 | |
| Deletion | Chr6 | 25917019 | 25917020 | 1 | |
| Deletion | Chr12 | 26015292 | 26015293 | 1 | |
| Deletion | Chr3 | 26017376 | 26017381 | 5 | |
| Deletion | Chr5 | 26394143 | 26394147 | 4 | |
| Deletion | Chr7 | 27162943 | 27162974 | 31 | LOC_Os07g45530 |
| Deletion | Chr1 | 29493753 | 29494009 | 256 |
Insertions: 7
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 13946854 | 14963285 | LOC_Os05g25720 |
| Inversion | Chr5 | 14153059 | 14963288 | 2 |
| Inversion | Chr8 | 17459060 | 17938751 | |
| Inversion | Chr12 | 24893664 | 24952412 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 25649125 | Chr2 | 6043437 | 2 |
