Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1251-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1251-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 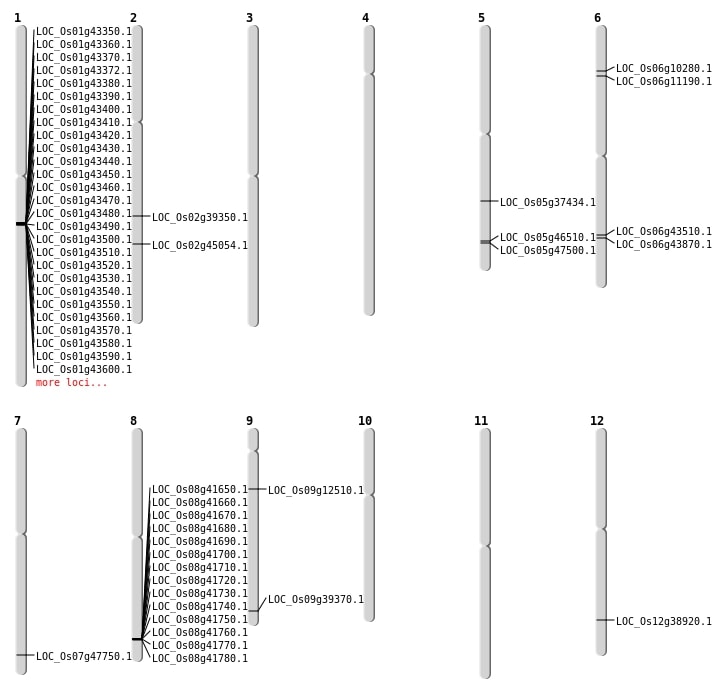
|
| Variant Information |
|---|
Single base substitutions: 24
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31311517 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g54420 |
| SBS | Chr1 | 37566180 | C-T | HOMO | UTR_5_PRIME | |
| SBS | Chr10 | 20340670 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 50859 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 9608324 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 10175062 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 16760003 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 18175144 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 26736800 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 21884577 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 27991797 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 26939826 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 681177 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 1718099 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 7337531 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 2786099 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 28532432 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g47750 |
| SBS | Chr7 | 28532433 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g47750 |
| SBS | Chr7 | 8486201 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 10230804 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 11104408 | A-G | HET | INTRON | |
| SBS | Chr8 | 11499787 | C-T | HET | INTRON | |
| SBS | Chr8 | 26856065 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7172869 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g12510 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 808327 | 808329 | 2 | |
| Deletion | Chr1 | 2560793 | 2560800 | 7 | |
| Deletion | Chr12 | 2878833 | 2878836 | 3 | |
| Deletion | Chr8 | 4285134 | 4285142 | 8 | |
| Deletion | Chr5 | 5614929 | 5614937 | 8 | |
| Deletion | Chr7 | 5834043 | 5834273 | 230 | |
| Deletion | Chr9 | 6090089 | 6090097 | 8 | |
| Deletion | Chr3 | 6418773 | 6418778 | 5 | |
| Deletion | Chr11 | 8428675 | 8428688 | 13 | |
| Deletion | Chr3 | 10798497 | 10798503 | 6 | |
| Deletion | Chr3 | 14008486 | 14008491 | 5 | |
| Deletion | Chr5 | 17948769 | 17948776 | 7 | |
| Deletion | Chr2 | 19090441 | 19090450 | 9 | |
| Deletion | Chr2 | 19800568 | 19800569 | 1 | |
| Deletion | Chr7 | 20785243 | 20785248 | 5 | |
| Deletion | Chr3 | 21449834 | 21449846 | 12 | |
| Deletion | Chr5 | 21906147 | 21906156 | 9 | LOC_Os05g37434 |
| Deletion | Chr1 | 22561864 | 22561866 | 2 | |
| Deletion | Chr9 | 22640046 | 22640048 | 2 | LOC_Os09g39370 |
| Deletion | Chr2 | 23750246 | 23750252 | 6 | LOC_Os02g39350 |
| Deletion | Chr1 | 24781001 | 25196000 | 415000 | 59 |
| Deletion | Chr1 | 25203001 | 25421000 | 218000 | 30 |
| Deletion | Chr8 | 26301001 | 26384000 | 83000 | 14 |
| Deletion | Chr12 | 26318018 | 26318026 | 8 | |
| Deletion | Chr5 | 26888877 | 26888904 | 27 | |
| Deletion | Chr2 | 27303375 | 27327355 | 23980 | LOC_Os02g45054 |
| Deletion | Chr5 | 27460576 | 27460582 | 6 | |
| Deletion | Chr1 | 29647835 | 29647854 | 19 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 26229922 | 26229923 | 2 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 5284966 | 5876342 | 2 |
| Inversion | Chr6 | 26158815 | 26876798 | LOC_Os06g43510 |
| Inversion | Chr6 | 26158827 | 26423738 | 2 |
| Inversion | Chr8 | 26301261 | 26834480 | LOC_Os08g41650 |
| Inversion | Chr8 | 26384030 | 26834483 | LOC_Os08g41780 |
| Inversion | Chr5 | 26939669 | 27212069 | LOC_Os05g47500 |
| Inversion | Chr5 | 26942091 | 27212081 | 2 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 20459693 | Chr6 | 26876803 | |
| Translocation | Chr8 | 20502339 | Chr6 | 26423739 | LOC_Os06g43870 |
| Translocation | Chr12 | 23933509 | Chr2 | 27303808 | LOC_Os12g38920 |
| Translocation | Chr12 | 23933543 | Chr2 | 27303439 | LOC_Os12g38920 |
| Translocation | Chr5 | 26939676 | Chr1 | 29927280 | |
| Translocation | Chr5 | 26942066 | Chr1 | 29927278 | LOC_Os05g46510 |
