Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1252-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1252-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 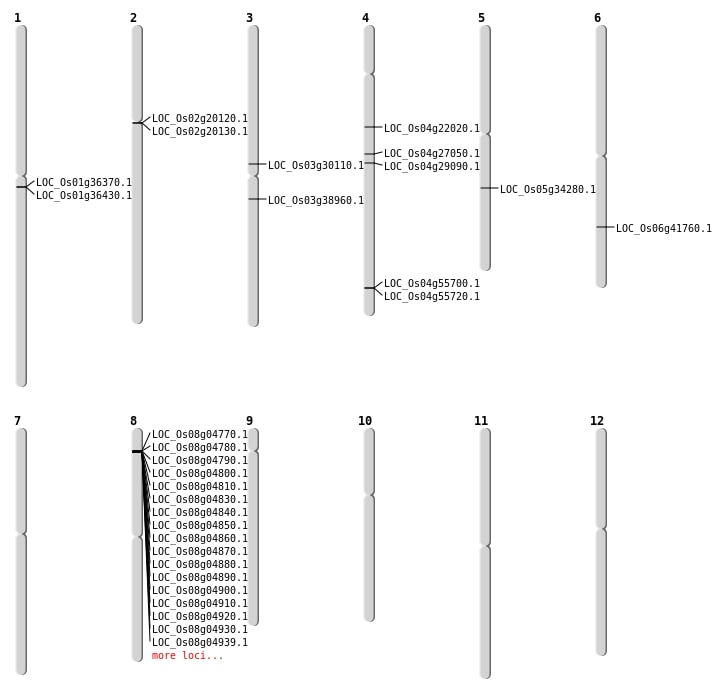
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20175389 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g36370 |
| SBS | Chr1 | 20186724 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20225314 | A-T | HET | STOP_GAINED | LOC_Os01g36430 |
| SBS | Chr1 | 25273995 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 3133114 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 3803793 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 5585468 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 12205360 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 2084016 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 6760238 | G-C | HET | INTRON | |
| SBS | Chr10 | 8989156 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 8989157 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14308109 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8422821 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 8687255 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 19540293 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 22122921 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 26929474 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 10780712 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 12473906 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22020 |
| SBS | Chr4 | 13144745 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 19309449 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 14009853 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 19033554 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 25877289 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 27674001 | C-T | HET | INTRON | |
| SBS | Chr6 | 11249126 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 12192611 | G-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 22296456 | C-T | HET | INTRON | |
| SBS | Chr6 | 29574444 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 2109101 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 22106116 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 854952 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 855458 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 5719991 | T-A | HOMO | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1445290 | 1445308 | 18 | |
| Deletion | Chr8 | 2379001 | 2563000 | 184000 | 21 |
| Deletion | Chr8 | 2611826 | 2611829 | 3 | LOC_Os08g05080 |
| Deletion | Chr8 | 3819047 | 3819531 | 484 | LOC_Os08g06840 |
| Deletion | Chr5 | 6303375 | 6303382 | 7 | |
| Deletion | Chr11 | 9074872 | 9074873 | 1 | |
| Deletion | Chr8 | 9375030 | 9375031 | 1 | |
| Deletion | Chr5 | 9986080 | 9986084 | 4 | |
| Deletion | Chr9 | 13309337 | 13309339 | 2 | |
| Deletion | Chr1 | 19717531 | 19717532 | 1 | |
| Deletion | Chr5 | 20289355 | 20289358 | 3 | LOC_Os05g34280 |
| Deletion | Chr12 | 21247509 | 21247511 | 2 | |
| Deletion | Chr6 | 22538612 | 22538616 | 4 | |
| Deletion | Chr8 | 23124491 | 23124497 | 6 | |
| Deletion | Chr5 | 23261268 | 23261282 | 14 | |
| Deletion | Chr5 | 23596143 | 23596144 | 1 | |
| Deletion | Chr6 | 25039092 | 25039098 | 6 | LOC_Os06g41760 |
| Deletion | Chr5 | 26184226 | 26184227 | 1 | |
| Deletion | Chr1 | 30215308 | 30215317 | 9 | |
| Deletion | Chr1 | 41570335 | 41570368 | 33 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 26576421 | 26576421 | 1 | |
| Insertion | Chr10 | 18111944 | 18111944 | 1 | |
| Insertion | Chr11 | 19345089 | 19345089 | 1 | |
| Insertion | Chr4 | 15999793 | 15999799 | 7 | LOC_Os04g27050 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 2563014 | 5568910 | |
| Inversion | Chr2 | 11854861 | 11860925 | 2 |
| Inversion | Chr3 | 17182701 | 26932425 | LOC_Os03g30110 |
| Inversion | Chr4 | 17260025 | 33168792 | 2 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2378619 | Chr3 | 6146750 | |
| Translocation | Chr8 | 2378700 | Chr4 | 33150847 | LOC_Os04g55700 |
| Translocation | Chr8 | 2378730 | Chr3 | 6146715 | |
| Translocation | Chr8 | 2562967 | Chr3 | 21639947 | LOC_Os03g38960 |
| Translocation | Chr8 | 5568864 | Chr3 | 21640100 | LOC_Os03g38960 |
| Translocation | Chr8 | 5568870 | Chr4 | 33153933 | |
| Translocation | Chr6 | 27804544 | Chr3 | 25799371 | |
| Translocation | Chr4 | 31134491 | Chr2 | 33189733 | |
| Translocation | Chr4 | 31134496 | Chr3 | 626738 | |
| Translocation | Chr4 | 31138297 | Chr3 | 626709 |
