Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1263-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1263-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 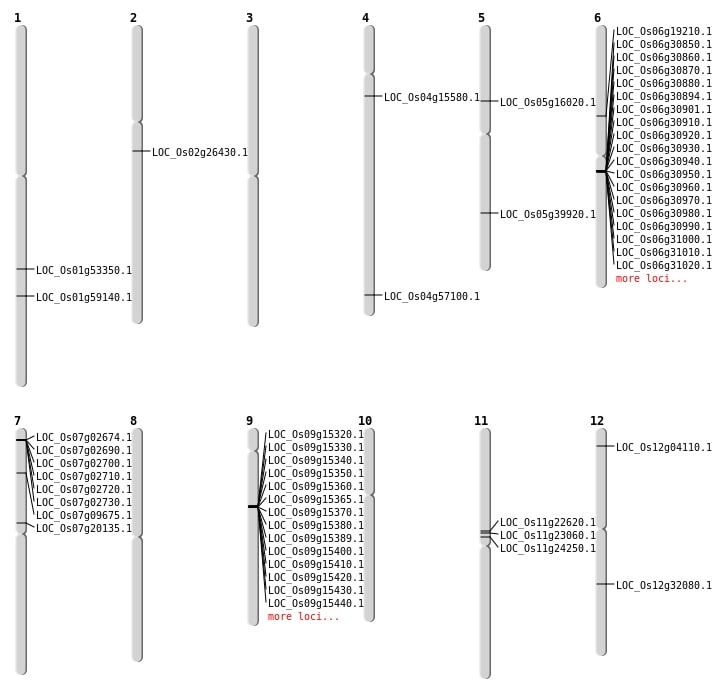
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30656359 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g53350 |
| SBS | Chr1 | 8582575 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 17396598 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 18933674 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 2302502 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 1676057 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 22896987 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 23470622 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 14740361 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 15417287 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 19354305 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g32080 |
| SBS | Chr12 | 20916991 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 12821490 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 15521605 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g26430 |
| SBS | Chr2 | 24964598 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 2653384 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 1979970 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 1979972 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 4703877 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5985379 | A-T | HET | INTRON | |
| SBS | Chr3 | 5985380 | A-T | HET | INTRON | |
| SBS | Chr4 | 19955200 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 26777291 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 13804115 | A-G | HET | INTRON | |
| SBS | Chr5 | 14520212 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr5 | 23458833 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g39920 |
| SBS | Chr5 | 9053231 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16020 |
| SBS | Chr6 | 15403490 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 21983343 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g37224 |
| SBS | Chr6 | 29942687 | C-A | HET | INTRON | |
| SBS | Chr6 | 30516435 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 17293394 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 19280151 | G-A | HET | INTRON | |
| SBS | Chr7 | 943188 | A-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 26784802 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 2789410 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 5206931 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 11890809 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 1938462 | G-A | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 765764 | 765765 | 1 | |
| Deletion | Chr7 | 969001 | 1000000 | 31000 | 6 |
| Deletion | Chr2 | 1718316 | 1718346 | 30 | |
| Deletion | Chr12 | 1721206 | 1721214 | 8 | LOC_Os12g04110 |
| Deletion | Chr7 | 5045703 | 5045725 | 22 | |
| Deletion | Chr7 | 5134629 | 5134636 | 7 | LOC_Os07g09675 |
| Deletion | Chr2 | 6232203 | 6232293 | 90 | |
| Deletion | Chr1 | 6679775 | 6679776 | 1 | |
| Deletion | Chr2 | 8419535 | 8419545 | 10 | |
| Deletion | Chr9 | 9356001 | 9879000 | 523000 | 89 |
| Deletion | Chr6 | 10934619 | 10934628 | 9 | LOC_Os06g19210 |
| Deletion | Chr4 | 12592457 | 12592469 | 12 | |
| Deletion | Chr9 | 14762769 | 14762778 | 9 | |
| Deletion | Chr6 | 17903001 | 18688000 | 785000 | 120 |
| Deletion | Chr5 | 19345819 | 19345820 | 1 | |
| Deletion | Chr1 | 19672244 | 19672256 | 12 | |
| Deletion | Chr6 | 23828703 | 23828704 | 1 | |
| Deletion | Chr3 | 27950701 | 27950728 | 27 | |
| Deletion | Chr4 | 34028322 | 34028334 | 12 | LOC_Os04g57100 |
| Deletion | Chr1 | 34164037 | 34164059 | 22 | LOC_Os01g59140 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 11615134 | 11615135 | 2 | |
| Insertion | Chr12 | 13506120 | 13506120 | 1 | |
| Insertion | Chr12 | 8869031 | 8869032 | 2 | |
| Insertion | Chr2 | 33978813 | 33978813 | 1 | |
| Insertion | Chr4 | 8474264 | 8474264 | 1 | LOC_Os04g15580 |
| Insertion | Chr7 | 11629690 | 11629703 | 14 | LOC_Os07g20135 |
| Insertion | Chr7 | 22241223 | 22241224 | 2 | |
| Insertion | Chr7 | 24000215 | 24000215 | 1 | |
| Insertion | Chr8 | 23181519 | 23181520 | 2 | |
| Insertion | Chr9 | 22681727 | 22681727 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 943070 | 999922 | |
| Inversion | Chr11 | 13002824 | 13814167 | LOC_Os11g24250 |
| Inversion | Chr11 | 13009569 | 13264292 | 2 |
| Inversion | Chr6 | 17887723 | 17892718 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 999659 | Chr1 | 21739882 | |
| Translocation | Chr7 | 999872 | Chr6 | 19323240 | |
| Translocation | Chr7 | 999901 | Chr1 | 21739924 | |
| Translocation | Chr6 | 19323241 | Chr1 | 21739883 | |
| Translocation | Chr6 | 19323518 | Chr1 | 21739872 | |
| Translocation | Chr5 | 25539360 | Chr1 | 40623992 |
