Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1264-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1264-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 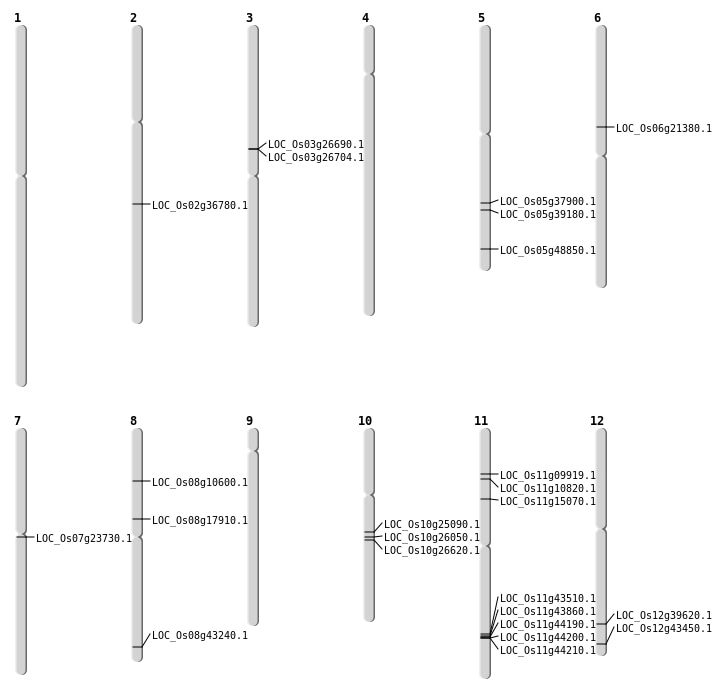
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29892130 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 38435424 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 38726659 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 43306 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 12926467 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g25090 |
| SBS | Chr10 | 3470004 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 15690487 | A-C | HET | INTRON | |
| SBS | Chr11 | 5969174 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g10820 |
| SBS | Chr12 | 22858418 | G-A | HET | INTRON | |
| SBS | Chr12 | 24420049 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 22604160 | C-T | HET | INTRON | |
| SBS | Chr2 | 26254385 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28954704 | G-A | HET | INTRON | |
| SBS | Chr2 | 31900357 | C-T | HET | INTRON | |
| SBS | Chr3 | 17085702 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 24739053 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 34352986 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 34832667 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 36311996 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 1159321 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 6527339 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 81924 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 9458535 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 9458536 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 10587606 | A-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 12672722 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17652576 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 19953420 | T-A | HET | INTRON | |
| SBS | Chr6 | 26770456 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 12464078 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 4718229 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 8437178 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 3643535 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 13805119 | C-G | HOMO | INTRON |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1961917 | 1961922 | 5 | |
| Deletion | Chr11 | 5311300 | 5311302 | 2 | LOC_Os11g09919 |
| Deletion | Chr8 | 6240443 | 6240446 | 3 | LOC_Os08g10600 |
| Deletion | Chr10 | 6988363 | 6988377 | 14 | |
| Deletion | Chr6 | 12349720 | 12349738 | 18 | LOC_Os06g21380 |
| Deletion | Chr8 | 15675057 | 15675069 | 12 | |
| Deletion | Chr9 | 18032629 | 18032632 | 3 | |
| Deletion | Chr1 | 21426733 | 21426735 | 2 | |
| Deletion | Chr3 | 22141082 | 22141091 | 9 | |
| Deletion | Chr2 | 22180439 | 22180444 | 5 | LOC_Os02g36780 |
| Deletion | Chr12 | 24454232 | 24455372 | 1140 | LOC_Os12g39620 |
| Deletion | Chr11 | 24647169 | 24647176 | 7 | |
| Deletion | Chr4 | 25924551 | 25924553 | 2 | |
| Deletion | Chr11 | 26689001 | 26704000 | 15000 | 3 |
| Deletion | Chr4 | 26907442 | 26907446 | 4 | |
| Deletion | Chr12 | 26960994 | 26961002 | 8 | LOC_Os12g43450 |
| Deletion | Chr8 | 27332903 | 27332909 | 6 | LOC_Os08g43240 |
| Deletion | Chr5 | 28003625 | 28003640 | 15 | LOC_Os05g48850 |
| Deletion | Chr1 | 38386150 | 38386151 | 1 |
Insertions: 8
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 13494081 | 13884607 | 2 |
| Inversion | Chr5 | 22210360 | 22977544 | 2 |
| Inversion | Chr5 | 24870612 | 24992151 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 8483266 | Chr2 | 30163186 | LOC_Os11g15070 |
| Translocation | Chr8 | 9573477 | Chr5 | 22210385 | LOC_Os05g37900 |
| Translocation | Chr7 | 10476836 | Chr1 | 1512463 | LOC_Os01g03660 |
| Translocation | Chr8 | 10991580 | Chr3 | 15245742 | 2 |
| Translocation | Chr5 | 14705823 | Chr2 | 12182619 | |
| Translocation | Chr3 | 15238126 | Chr2 | 3401782 | LOC_Os03g26690 |
| Translocation | Chr11 | 26265710 | Chr7 | 13389429 | 2 |
| Translocation | Chr11 | 26492320 | Chr2 | 30163179 | LOC_Os11g43860 |
