Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1265-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1265-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 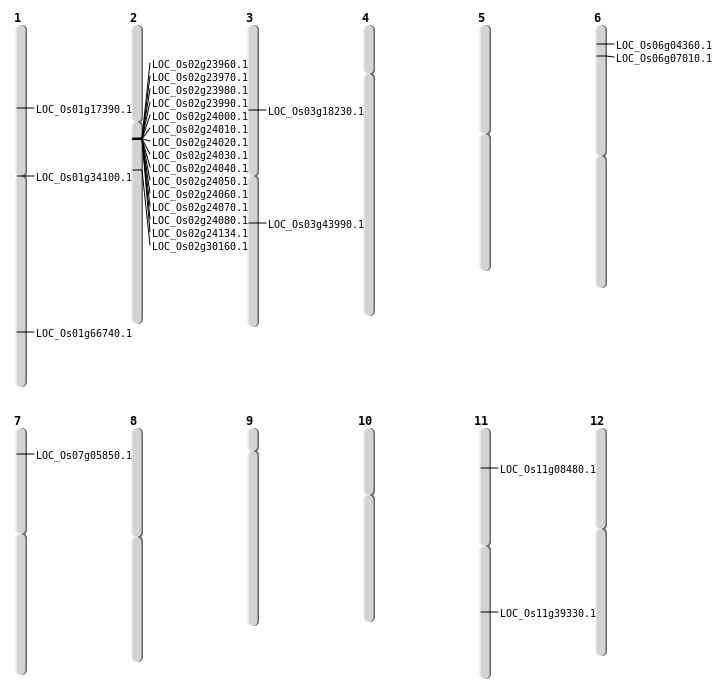
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21103802 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 27925054 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 30016585 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 34311847 | T-G | HET | INTRON | |
| SBS | Chr1 | 9900514 | C-T | HET | INTRON | |
| SBS | Chr11 | 15410703 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 4481847 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08480 |
| SBS | Chr12 | 20297810 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 24045276 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr2 | 14386950 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 17919865 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g30160 |
| SBS | Chr2 | 20513106 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 31842719 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 33108888 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 8807055 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 1723861 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 23843401 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 24407220 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 26696178 | T-C | HET | INTRON | |
| SBS | Chr5 | 27516401 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 23289365 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 23895900 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 28593499 | G-C | HET | INTRON | |
| SBS | Chr6 | 3324974 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g07010 |
| SBS | Chr6 | 3343837 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 27576931 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 20257874 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 4493240 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 4931363 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 6631333 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 13342129 | G-T | HOMO | INTRON | |
| SBS | Chr9 | 16149906 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9102812 | G-T | HOMO | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 113657 | 113663 | 6 | LOC_Os03g01160 |
| Deletion | Chr7 | 2813726 | 2813730 | 4 | LOC_Os07g05850 |
| Deletion | Chr8 | 3221803 | 3221827 | 24 | |
| Deletion | Chr4 | 7190225 | 7190229 | 4 | |
| Deletion | Chr10 | 9313175 | 9313177 | 2 | |
| Deletion | Chr12 | 12438652 | 12438659 | 7 | |
| Deletion | Chr6 | 13845075 | 13845106 | 31 | |
| Deletion | Chr2 | 13878001 | 13952000 | 74000 | 13 |
| Deletion | Chr2 | 13967001 | 13980000 | 13000 | LOC_Os02g24134 |
| Deletion | Chr5 | 15947975 | 15947980 | 5 | |
| Deletion | Chr11 | 18003501 | 18003503 | 2 | |
| Deletion | Chr9 | 18815816 | 18815825 | 9 | |
| Deletion | Chr5 | 19424667 | 19424668 | 1 | |
| Deletion | Chr10 | 20122801 | 20122820 | 19 | |
| Deletion | Chr2 | 21946243 | 21946332 | 89 | |
| Deletion | Chr2 | 25243357 | 25243364 | 7 | |
| Deletion | Chr6 | 26149694 | 26149697 | 3 | |
| Deletion | Chr4 | 32065013 | 32065016 | 3 | |
| Deletion | Chr1 | 38761844 | 38761847 | 3 | LOC_Os01g66740 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18781253 | 18781253 | 1 | LOC_Os01g34100 |
| Insertion | Chr11 | 23416708 | 23416708 | 1 | LOC_Os11g39330 |
| Insertion | Chr11 | 27585620 | 27585621 | 2 | |
| Insertion | Chr2 | 18089824 | 18089825 | 2 | |
| Insertion | Chr3 | 10224923 | 10224924 | 2 | LOC_Os03g18230 |
| Insertion | Chr5 | 12107831 | 12107831 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 1854575 | 3326332 | LOC_Os06g07010 |
| Inversion | Chr6 | 1858584 | 3326308 | 2 |
| Inversion | Chr2 | 13877755 | 14206589 | |
| Inversion | Chr2 | 13979938 | 14206594 | LOC_Os02g24134 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1854503 | Chr4 | 22146747 | |
| Translocation | Chr6 | 3326292 | Chr4 | 22146572 | LOC_Os06g07010 |
| Translocation | Chr11 | 13216944 | Chr10 | 18828727 | |
| Translocation | Chr3 | 24696564 | Chr1 | 10007551 | LOC_Os01g17390 |
| Translocation | Chr3 | 24702160 | Chr2 | 21946245 | LOC_Os03g43990 |
| Translocation | Chr3 | 24702200 | Chr1 | 10007598 | 2 |
