Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1266-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1266-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 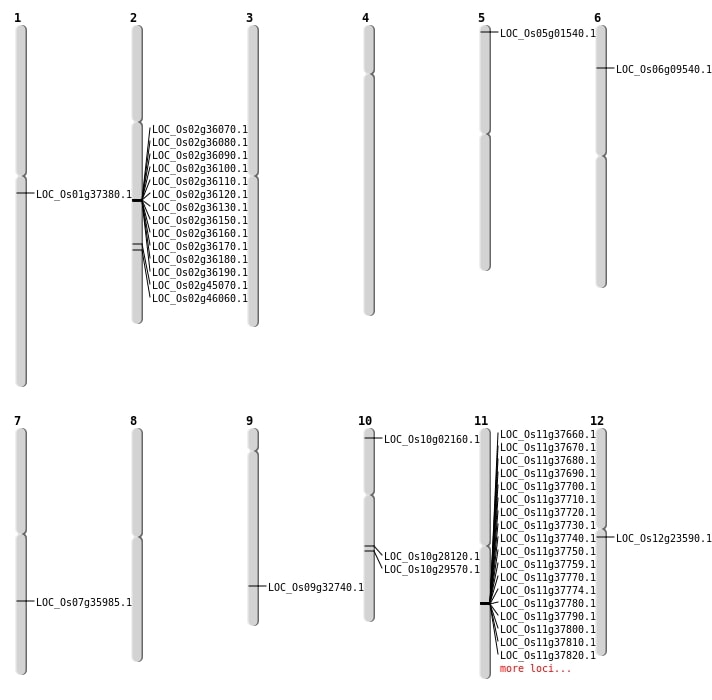
|
| Variant Information |
|---|
Single base substitutions: 23
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25294021 | T-G | HOMO | INTRON | |
| SBS | Chr1 | 27328086 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 27328089 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 4805437 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11683368 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 14602857 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g28120 |
| SBS | Chr10 | 7318949 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 15764663 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 20896531 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3416331 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 21498334 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 23031047 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 5776600 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 5776605 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 5776613 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 9405193 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 11296652 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 23532438 | C-A | HET | INTRON | |
| SBS | Chr7 | 27750671 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 10012170 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 17612235 | T-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 21360769 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 8810960 | A-T | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 736656 | 736657 | 1 | LOC_Os10g02160 |
| Deletion | Chr2 | 3236458 | 3236459 | 1 | |
| Deletion | Chr1 | 3709021 | 3709037 | 16 | |
| Deletion | Chr6 | 4854365 | 4854369 | 4 | LOC_Os06g09540 |
| Deletion | Chr10 | 7902863 | 7902865 | 2 | |
| Deletion | Chr12 | 11535447 | 11535455 | 8 | |
| Deletion | Chr9 | 17383865 | 17383903 | 38 | |
| Deletion | Chr4 | 18974573 | 18974582 | 9 | |
| Deletion | Chr9 | 19524599 | 19524605 | 6 | LOC_Os09g32740 |
| Deletion | Chr1 | 20872097 | 20872108 | 11 | LOC_Os01g37380 |
| Deletion | Chr7 | 21526697 | 21527031 | 334 | LOC_Os07g35985 |
| Deletion | Chr2 | 21682794 | 21816766 | 133972 | 13 |
| Deletion | Chr10 | 21992972 | 21992973 | 1 | |
| Deletion | Chr11 | 22223001 | 22525000 | 302000 | 36 |
| Deletion | Chr11 | 22530001 | 22562000 | 32000 | 4 |
| Deletion | Chr11 | 22564001 | 23196000 | 632000 | 89 |
| Deletion | Chr7 | 24929715 | 24929725 | 10 | |
| Deletion | Chr11 | 26483951 | 26483953 | 2 | |
| Deletion | Chr4 | 28287182 | 28287183 | 1 | |
| Deletion | Chr1 | 35488370 | 35488371 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 13390402 | 13390403 | 2 | |
| Insertion | Chr3 | 17333768 | 17333768 | 1 | |
| Insertion | Chr7 | 736926 | 736926 | 1 | |
| Insertion | Chr8 | 26815767 | 26815768 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 21362594 | 21498394 | |
| Inversion | Chr2 | 21498433 | 21682804 | |
| Inversion | Chr2 | 27334120 | 28068845 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 13372018 | Chr10 | 16479598 | LOC_Os12g23590 |
| Translocation | Chr12 | 13386894 | Chr10 | 15370828 | LOC_Os10g29570 |
| Translocation | Chr7 | 21527010 | Chr2 | 27334123 | 2 |
| Translocation | Chr11 | 23187874 | Chr5 | 317260 | LOC_Os05g01540 |
