Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1270-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1270-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 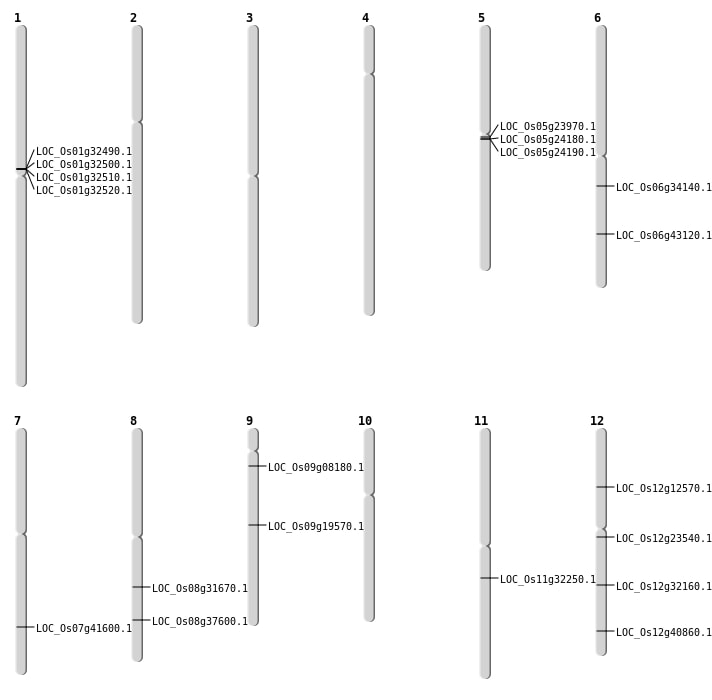
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17090217 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 17816344 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 17816345 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 29675646 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 15685641 | C-T | HET | INTRON | |
| SBS | Chr11 | 19056214 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g32250 |
| SBS | Chr11 | 8735013 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 12380388 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 13324732 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23540 |
| SBS | Chr12 | 19407658 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g32160 |
| SBS | Chr12 | 291601 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 5288221 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 26304188 | T-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 17752493 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 22259667 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 24315106 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 24635692 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 24669647 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14360575 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 11489228 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 15677004 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 9936315 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 14635121 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15728444 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17419281 | G-A | HET | INTRON | |
| SBS | Chr6 | 19892174 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g34140 |
| SBS | Chr7 | 14930976 | C-A | HOMO | INTRON | |
| SBS | Chr7 | 23995263 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 24982855 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 14411674 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 18170424 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 5697275 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 9335198 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 11709481 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19570 |
| SBS | Chr9 | 4241381 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g08180 |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr12 | 6914779 | 6914791 | 12 | LOC_Os12g12570 |
| Deletion | Chr8 | 10393235 | 10393236 | 1 | |
| Deletion | Chr5 | 13802270 | 13802280 | 10 | LOC_Os05g23970 |
| Deletion | Chr5 | 13961589 | 13978397 | 16808 | 2 |
| Deletion | Chr1 | 17816245 | 17838782 | 22537 | 4 |
| Deletion | Chr1 | 19317713 | 19317720 | 7 | |
| Deletion | Chr8 | 19635014 | 19635020 | 6 | LOC_Os08g31670 |
| Deletion | Chr3 | 24315122 | 24315123 | 1 | |
| Deletion | Chr12 | 25301228 | 25301242 | 14 | LOC_Os12g40860 |
| Deletion | Chr6 | 26162689 | 26162690 | 1 | |
| Deletion | Chr8 | 27491102 | 27491107 | 5 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23930692 | 23930693 | 2 | |
| Insertion | Chr1 | 34967776 | 34967776 | 1 | |
| Insertion | Chr11 | 17018891 | 17018892 | 2 | |
| Insertion | Chr12 | 22090334 | 22090335 | 2 | |
| Insertion | Chr5 | 14161161 | 14161170 | 10 | |
| Insertion | Chr7 | 23010388 | 23010389 | 2 | |
| Insertion | Chr7 | 24934517 | 24934519 | 3 | LOC_Os07g41600 |
| Insertion | Chr9 | 7766568 | 7766568 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 14654899 | 14662855 | |
| Inversion | Chr11 | 20865403 | 28679730 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 13969188 | Chr8 | 23824948 | LOC_Os08g37600 |
| Translocation | Chr6 | 25918653 | Chr1 | 17816287 | LOC_Os06g43120 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr1 | 17808683 | 17838778 | 30095 |
