Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1278-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1278-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 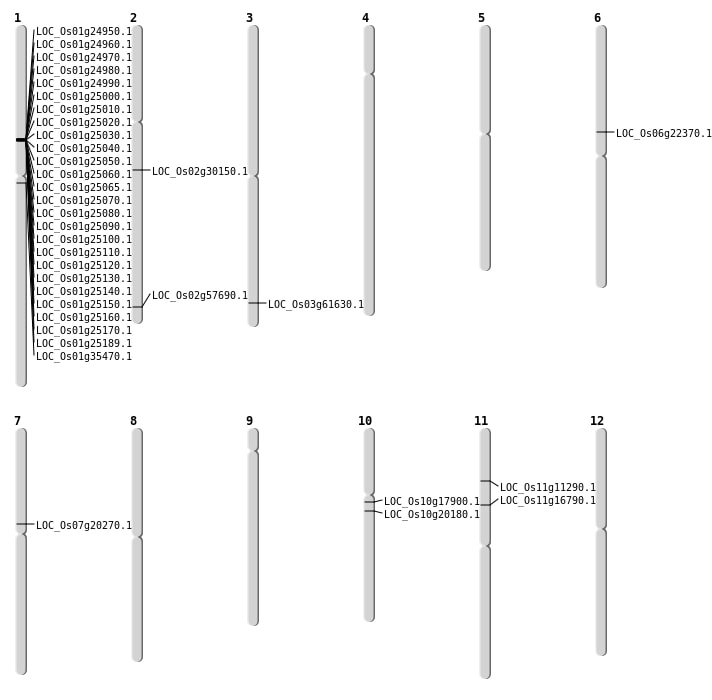
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11608707 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 13236707 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 18502084 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 19626536 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g35470 |
| SBS | Chr1 | 2958775 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 34209005 | T-A | HET | INTRON | |
| SBS | Chr1 | 34209006 | T-A | HET | INTRON | |
| SBS | Chr1 | 41043808 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 5431653 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 5995170 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 23053497 | A-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 23053498 | C-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 9048183 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g17900 |
| SBS | Chr11 | 6250398 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g11290 |
| SBS | Chr11 | 9316026 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g16790 |
| SBS | Chr11 | 9728930 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9728932 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9728935 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11207680 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 17846224 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 23011274 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 26875728 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 8168331 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 9030948 | A-C | HET | INTRON | |
| SBS | Chr2 | 17213996 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 17214370 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 23561874 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 30190365 | C-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr2 | 6575279 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 16792857 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 21145250 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 25078048 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 4839864 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 12999371 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 14875767 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 26293214 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 10714491 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 1141702 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 13594042 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 5108442 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 12988236 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g22370 |
| SBS | Chr6 | 13176802 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 13827410 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 17185842 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 27669943 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 16695555 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 15340786 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4886230 | A-G | HOMO | INTRON | |
| SBS | Chr9 | 5257029 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 8218304 | C-G | HOMO | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 219125 | 219128 | 3 | |
| Deletion | Chr9 | 297889 | 297899 | 10 | |
| Deletion | Chr6 | 621173 | 621178 | 5 | |
| Deletion | Chr7 | 6502471 | 6502476 | 5 | |
| Deletion | Chr8 | 6788458 | 6788481 | 23 | |
| Deletion | Chr3 | 7454642 | 7454643 | 1 | |
| Deletion | Chr7 | 7522401 | 7522406 | 5 | |
| Deletion | Chr4 | 8174288 | 8174299 | 11 | |
| Deletion | Chr10 | 10132966 | 10132977 | 11 | LOC_Os10g20180 |
| Deletion | Chr9 | 10162636 | 10162637 | 1 | |
| Deletion | Chr8 | 10954773 | 10954776 | 3 | |
| Deletion | Chr7 | 11687148 | 11687151 | 3 | LOC_Os07g20270 |
| Deletion | Chr11 | 11699922 | 11699923 | 1 | |
| Deletion | Chr11 | 11803747 | 11803764 | 17 | |
| Deletion | Chr12 | 12758354 | 12758368 | 14 | |
| Deletion | Chr1 | 14062934 | 14236935 | 174001 | 25 |
| Deletion | Chr8 | 17551901 | 17551941 | 40 | |
| Deletion | Chr2 | 17911281 | 17911290 | 9 | LOC_Os02g30150 |
| Deletion | Chr4 | 19128716 | 19128724 | 8 | |
| Deletion | Chr8 | 21270224 | 21270226 | 2 | |
| Deletion | Chr8 | 21947344 | 21947350 | 6 | |
| Deletion | Chr3 | 23523199 | 23523206 | 7 | |
| Deletion | Chr6 | 24088460 | 24088464 | 4 | |
| Deletion | Chr3 | 24160296 | 24160297 | 1 | |
| Deletion | Chr1 | 24358226 | 24358227 | 1 | |
| Deletion | Chr6 | 24521563 | 24521573 | 10 | |
| Deletion | Chr7 | 27249307 | 27249312 | 5 | |
| Deletion | Chr2 | 31168691 | 31168694 | 3 | |
| Deletion | Chr2 | 32453196 | 32453204 | 8 | |
| Deletion | Chr2 | 32901213 | 32901221 | 8 | |
| Deletion | Chr3 | 34941315 | 34941325 | 10 | LOC_Os03g61630 |
| Deletion | Chr2 | 35327026 | 35330134 | 3108 | LOC_Os02g57690 |
| Deletion | Chr1 | 38914084 | 38914093 | 9 |
Insertions: 5
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2324484 | Chr2 | 10167366 | |
| Translocation | Chr6 | 14778297 | Chr3 | 5107995 | |
| Translocation | Chr6 | 14779253 | Chr3 | 5107974 | |
| Translocation | Chr11 | 22349561 | Chr3 | 5107950 | |
| Translocation | Chr3 | 29049448 | Chr2 | 35330132 | LOC_Os02g57690 |
| Translocation | Chr3 | 29049459 | Chr2 | 35327035 | LOC_Os02g57690 |
