Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1280-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1280-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 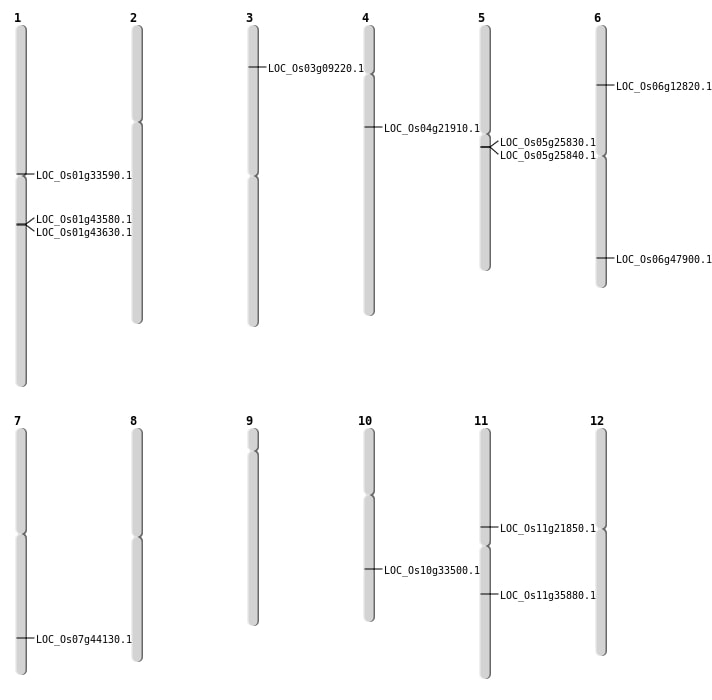
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14797339 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 18087082 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 18485249 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g33590 |
| SBS | Chr1 | 39645830 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 5969967 | C-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 20880511 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 13594143 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 21074471 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g35880 |
| SBS | Chr11 | 8072193 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1532199 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 15077683 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 4702225 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 9590433 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 20995448 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 27419608 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 28216393 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 4818618 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 4819636 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g09220 |
| SBS | Chr3 | 6763361 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 10934008 | G-A | HET | INTRON | |
| SBS | Chr4 | 16412181 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 2163939 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 2211002 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 22843086 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 2894290 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 19187046 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 13617497 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 21157378 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 4587335 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 6327598 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 9047596 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 14509175 | T-C | HET | INTRON | |
| SBS | Chr7 | 9342439 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 23856392 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 6031419 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 9422088 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 10913480 | T-C | HOMO | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 7006384 | 7006387 | 3 | LOC_Os06g12820 |
| Deletion | Chr4 | 12406442 | 12406446 | 4 | LOC_Os04g21910 |
| Deletion | Chr11 | 12550344 | 12550349 | 5 | LOC_Os11g21850 |
| Deletion | Chr2 | 12740683 | 12740687 | 4 | |
| Deletion | Chr6 | 13395663 | 13395669 | 6 | |
| Deletion | Chr11 | 13652685 | 13652687 | 2 | |
| Deletion | Chr11 | 14992218 | 14992222 | 4 | |
| Deletion | Chr5 | 15036001 | 15045000 | 9000 | 2 |
| Deletion | Chr5 | 16366390 | 16366396 | 6 | |
| Deletion | Chr1 | 16698057 | 16698059 | 2 | |
| Deletion | Chr3 | 17902263 | 17902264 | 1 | |
| Deletion | Chr11 | 24801724 | 24801728 | 4 | |
| Deletion | Chr7 | 25877718 | 25877723 | 5 | |
| Deletion | Chr7 | 25886149 | 25886150 | 1 | |
| Deletion | Chr7 | 26376959 | 26376965 | 6 | LOC_Os07g44130 |
| Deletion | Chr7 | 27464635 | 27464642 | 7 | |
| Deletion | Chr6 | 28977821 | 28977827 | 6 | LOC_Os06g47900 |
| Deletion | Chr1 | 29344198 | 29344200 | 2 | |
| Deletion | Chr2 | 33524094 | 33524103 | 9 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 8582698 | 8582718 | 21 | |
| Insertion | Chr11 | 19595026 | 19595027 | 2 | |
| Insertion | Chr6 | 1309350 | 1309351 | 2 | |
| Insertion | Chr7 | 8045906 | 8045909 | 4 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 15036139 | 15254072 | LOC_Os05g25830 |
| Inversion | Chr5 | 15045162 | 15254107 | |
| Inversion | Chr5 | 28241560 | 28302174 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8523391 | Chr6 | 21524123 | |
| Translocation | Chr10 | 17654154 | Chr4 | 4446210 | LOC_Os10g33500 |
| Translocation | Chr2 | 33982013 | Chr1 | 24997041 | LOC_Os01g43630 |
| Translocation | Chr2 | 33982016 | Chr1 | 24958803 | LOC_Os01g43580 |
