Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1282-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1282-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 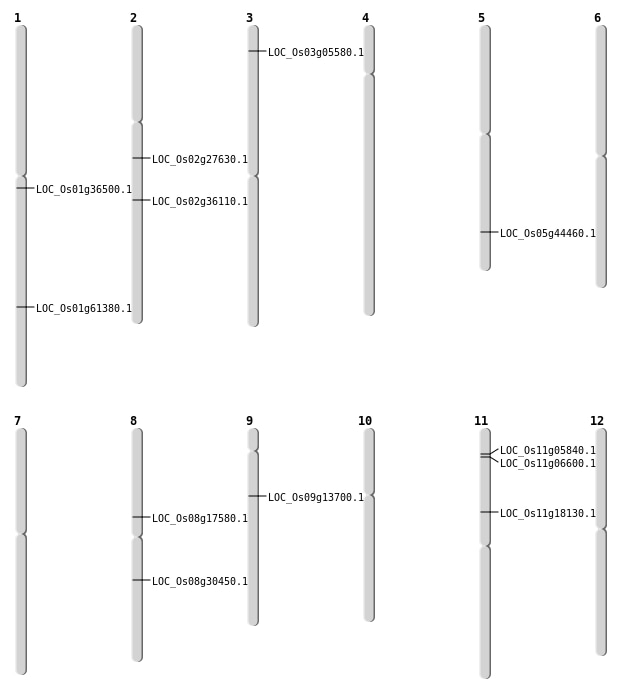
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24666887 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 28891196 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 31546841 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 31836183 | A-T | HET | INTRON | |
| SBS | Chr1 | 33187588 | A-T | HET | INTRON | |
| SBS | Chr1 | 8675325 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 17658548 | C-A | HET | INTRON | |
| SBS | Chr10 | 20768531 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr10 | 2410846 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5828868 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 9694198 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 10204655 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g18130 |
| SBS | Chr11 | 14602322 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 3188762 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g06600 |
| SBS | Chr11 | 6094394 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 17309110 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 23102114 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr12 | 5372592 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 11992131 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 16374949 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g27630 |
| SBS | Chr3 | 16676488 | G-T | HET | INTRON | |
| SBS | Chr3 | 18090985 | G-C | HET | INTRON | |
| SBS | Chr3 | 20624583 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 2793590 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g05580 |
| SBS | Chr3 | 32924155 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 10961282 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 12304932 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 16024197 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 17157330 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 17879767 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 24730672 | T-G | HET | INTRON | |
| SBS | Chr5 | 25878251 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44460 |
| SBS | Chr5 | 8489173 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 21283294 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 23495551 | G-A | HET | INTRON | |
| SBS | Chr6 | 23858296 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18511996 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 7379412 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 14432424 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 12947541 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 3248980 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 6982883 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8011795 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g13700 |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1210081 | 1210089 | 8 | |
| Deletion | Chr11 | 2717899 | 2717905 | 6 | LOC_Os11g05840 |
| Deletion | Chr3 | 2868949 | 2868951 | 2 | |
| Deletion | Chr11 | 3195954 | 3195971 | 17 | |
| Deletion | Chr2 | 3281041 | 3281042 | 1 | |
| Deletion | Chr11 | 3468433 | 3468437 | 4 | |
| Deletion | Chr1 | 3767779 | 3767780 | 1 | |
| Deletion | Chr2 | 4492662 | 4492663 | 1 | |
| Deletion | Chr12 | 5600633 | 5600655 | 22 | |
| Deletion | Chr5 | 7124822 | 7124823 | 1 | |
| Deletion | Chr11 | 8804580 | 8804584 | 4 | |
| Deletion | Chr2 | 11692862 | 11692874 | 12 | |
| Deletion | Chr5 | 12059061 | 12059066 | 5 | |
| Deletion | Chr9 | 12973540 | 12973541 | 1 | |
| Deletion | Chr9 | 16795687 | 16795693 | 6 | |
| Deletion | Chr6 | 17821244 | 17821256 | 12 | |
| Deletion | Chr7 | 18516391 | 18516398 | 7 | |
| Deletion | Chr8 | 18745209 | 18745215 | 6 | LOC_Os08g30450 |
| Deletion | Chr12 | 20979521 | 20979523 | 2 | |
| Deletion | Chr7 | 21478373 | 21478385 | 12 | |
| Deletion | Chr2 | 21740897 | 21740910 | 13 | LOC_Os02g36110 |
| Deletion | Chr12 | 22221347 | 22221350 | 3 | |
| Deletion | Chr3 | 23447569 | 23447573 | 4 | |
| Deletion | Chr5 | 25990512 | 25990520 | 8 | |
| Deletion | Chr2 | 26239410 | 26239419 | 9 | |
| Deletion | Chr11 | 28275725 | 28275795 | 70 | |
| Deletion | Chr4 | 34935143 | 34935144 | 1 | |
| Deletion | Chr1 | 35500839 | 35500848 | 9 | LOC_Os01g61380 |
| Deletion | Chr3 | 36141704 | 36141706 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2564497 | 2564498 | 2 | |
| Insertion | Chr12 | 22918377 | 22918377 | 1 | |
| Insertion | Chr6 | 26153097 | 26153099 | 3 |
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 7336063 | Chr1 | 20257176 | LOC_Os01g36500 |
| Translocation | Chr9 | 7641633 | Chr1 | 26359239 | |
| Translocation | Chr8 | 10754458 | Chr6 | 19260584 | LOC_Os08g17580 |
| Translocation | Chr8 | 11492916 | Chr2 | 9474097 | |
| Translocation | Chr11 | 28280603 | Chr3 | 7009451 |
