Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1285-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1285-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 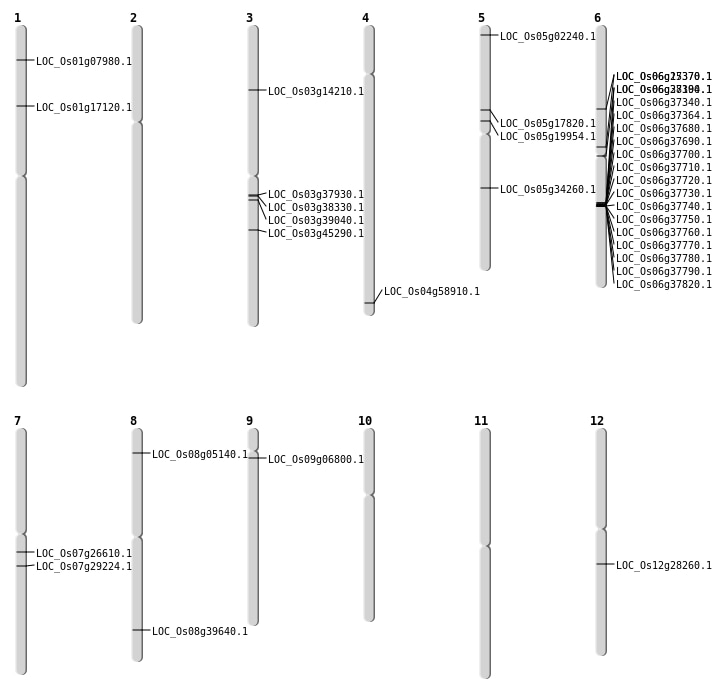
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12586442 | T-A | HOMO | INTRON | |
| SBS | Chr1 | 35866051 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 3860974 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g07980 |
| SBS | Chr1 | 4834090 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 18414388 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 15070632 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 19234518 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 25329773 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5708148 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 16703003 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g28260 |
| SBS | Chr12 | 17850606 | C-G | HOMO | INTRON | |
| SBS | Chr12 | 8716649 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 16854484 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 36311133 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 12905308 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 27147944 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 12345068 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 14460975 | C-A | HET | INTRON | |
| SBS | Chr5 | 20278696 | C-T | HET | STOP_GAINED | LOC_Os05g34260 |
| SBS | Chr5 | 20278697 | C-T | HET | STOP_GAINED | LOC_Os05g34260 |
| SBS | Chr5 | 20366210 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 10067600 | G-A | HET | STOP_GAINED | LOC_Os06g17370 |
| SBS | Chr6 | 14860784 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25370 |
| SBS | Chr6 | 22372655 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 9842724 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 15345061 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g26610 |
| SBS | Chr7 | 16745836 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 7175061 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 9206890 | A-G | HET | INTRON | |
| SBS | Chr8 | 11586558 | C-A | HOMO | INTRON | |
| SBS | Chr8 | 11642093 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 25102253 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g39640 |
| SBS | Chr9 | 3252028 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g06800 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 3436616 | 3436620 | 4 | |
| Deletion | Chr8 | 3544142 | 3544143 | 1 | |
| Deletion | Chr1 | 4018817 | 4018827 | 10 | |
| Deletion | Chr4 | 5329670 | 5329671 | 1 | |
| Deletion | Chr4 | 7107715 | 7107716 | 1 | |
| Deletion | Chr3 | 7707650 | 7707655 | 5 | LOC_Os03g14210 |
| Deletion | Chr2 | 7786063 | 7786072 | 9 | |
| Deletion | Chr6 | 9606170 | 9606174 | 4 | |
| Deletion | Chr1 | 9842039 | 9842050 | 11 | LOC_Os01g17120 |
| Deletion | Chr5 | 10249875 | 10249882 | 7 | LOC_Os05g17820 |
| Deletion | Chr5 | 11656218 | 11656224 | 6 | LOC_Os05g19954 |
| Deletion | Chr4 | 12048800 | 12048806 | 6 | |
| Deletion | Chr3 | 12388765 | 12388770 | 5 | |
| Deletion | Chr3 | 17419205 | 17419209 | 4 | |
| Deletion | Chr2 | 19868073 | 19868084 | 11 | |
| Deletion | Chr3 | 21071193 | 21071206 | 13 | |
| Deletion | Chr11 | 21676167 | 21676172 | 5 | |
| Deletion | Chr6 | 21962018 | 21962019 | 1 | |
| Deletion | Chr6 | 22070001 | 22086000 | 16000 | 2 |
| Deletion | Chr6 | 22328001 | 22369000 | 41000 | 12 |
| Deletion | Chr3 | 31479009 | 31479015 | 6 | |
| Deletion | Chr4 | 35039990 | 35040005 | 15 | LOC_Os04g58910 |
Insertions: 5
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 714805 | 715144 | LOC_Os05g02240 |
| Inversion | Chr6 | 16020883 | 16281530 | LOC_Os06g28194 |
| Inversion | Chr6 | 16020899 | 16281735 | LOC_Os06g28194 |
| Inversion | Chr3 | 21061133 | 21268643 | 2 |
| Inversion | Chr3 | 21297665 | 21646474 | |
| Inversion | Chr6 | 21669806 | 22263451 | |
| Inversion | Chr6 | 22022118 | 22378305 | 2 |
| Inversion | Chr6 | 22070311 | 22372858 | LOC_Os06g37340 |
| Inversion | Chr6 | 22263377 | 22327919 | LOC_Os06g37680 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2659188 | Chr3 | 26696327 | LOC_Os08g05140 |
| Translocation | Chr3 | 4463928 | Chr2 | 2674028 | |
| Translocation | Chr11 | 7272760 | Chr7 | 17129582 | LOC_Os07g29224 |
| Translocation | Chr11 | 11660361 | Chr10 | 5044332 | |
| Translocation | Chr12 | 13389110 | Chr7 | 19610118 | |
| Translocation | Chr6 | 19916319 | Chr3 | 25571191 | LOC_Os03g45290 |
| Translocation | Chr6 | 22070264 | Chr3 | 21297645 | LOC_Os06g37340 |
| Translocation | Chr6 | 22372803 | Chr3 | 21297397 | |
| Translocation | Chr4 | 27746232 | Chr3 | 16831606 |
Tandem duplications: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 21268652 | 21268652 | 0 | LOC_Os03g38330 |
| Tandem Duplication | Chr3 | 21702219 | 21702219 | 0 | LOC_Os03g39040 |
