Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1287-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1287-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 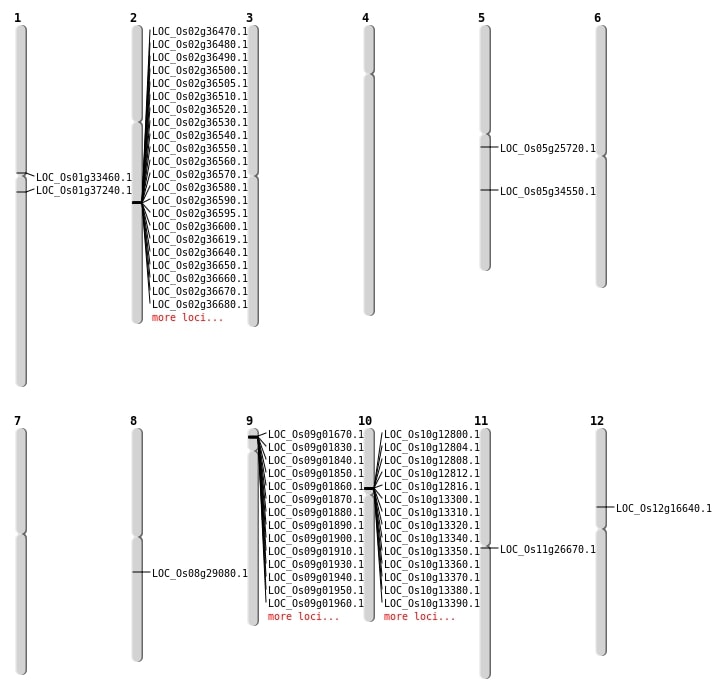
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12189269 | G-A | HET | INTRON | |
| SBS | Chr1 | 30822066 | A-G | HET | INTRON | |
| SBS | Chr1 | 40225892 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 40225895 | T-A | HOMO | INTRON | |
| SBS | Chr10 | 339069 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3753431 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 13090766 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 15275959 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26670 |
| SBS | Chr11 | 27382432 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 14850755 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 20565960 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 9529143 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16640 |
| SBS | Chr2 | 17024515 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 35282085 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 4052247 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr3 | 17656984 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 34577392 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 6910565 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 13341946 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 33910701 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 5156198 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 5519339 | G-A | HET | INTRON | |
| SBS | Chr5 | 12051264 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 16883702 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16883706 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 27542771 | A-C | HET | INTRON | |
| SBS | Chr6 | 12370057 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 14728388 | A-G | HET | INTRON | |
| SBS | Chr6 | 26357036 | T-A | HET | INTRON | |
| SBS | Chr7 | 16830023 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 22531424 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 3407109 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 492811 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8385420 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 17802107 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g29080 |
| SBS | Chr8 | 18635102 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 26987926 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 13303370 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g21980 |
| SBS | Chr9 | 16664748 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 17428018 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17428019 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g28660 |
| SBS | Chr9 | 20627322 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3568206 | C-T | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 569001 | 798000 | 229000 | 28 |
| Deletion | Chr8 | 620925 | 620936 | 11 | |
| Deletion | Chr3 | 4331007 | 4331010 | 3 | |
| Deletion | Chr2 | 5398468 | 5398469 | 1 | |
| Deletion | Chr8 | 6139172 | 6139182 | 10 | |
| Deletion | Chr10 | 7147001 | 7504000 | 357000 | 53 |
| Deletion | Chr11 | 8331018 | 8331031 | 13 | |
| Deletion | Chr5 | 14962089 | 14962090 | 1 | LOC_Os05g25720 |
| Deletion | Chr1 | 18088495 | 18088504 | 9 | |
| Deletion | Chr1 | 18430741 | 18430744 | 3 | LOC_Os01g33460 |
| Deletion | Chr1 | 20076536 | 20076538 | 2 | |
| Deletion | Chr5 | 20487498 | 20487499 | 1 | LOC_Os05g34550 |
| Deletion | Chr1 | 20798776 | 20798793 | 17 | LOC_Os01g37240 |
| Deletion | Chr2 | 22023001 | 22269000 | 246000 | 44 |
| Deletion | Chr2 | 22690707 | 22690720 | 13 | |
| Deletion | Chr9 | 22705324 | 22707105 | 1781 | LOC_Os09g39550 |
| Deletion | Chr12 | 24452200 | 24452204 | 4 | |
| Deletion | Chr2 | 25045872 | 25045889 | 17 | |
| Deletion | Chr4 | 27686905 | 27686911 | 6 | |
| Deletion | Chr2 | 34475602 | 34475603 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 8324723 | 8324723 | 1 | |
| Insertion | Chr4 | 312993 | 312994 | 2 | |
| Insertion | Chr7 | 29200801 | 29200801 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 483302 | 5903475 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 733680 | Chr8 | 4848745 | LOC_Os09g02030 |
| Translocation | Chr9 | 734526 | Chr8 | 4848707 | LOC_Os09g02030 |
| Translocation | Chr9 | 5944712 | Chr3 | 31777524 | |
| Translocation | Chr9 | 5950286 | Chr3 | 31777482 | |
| Translocation | Chr7 | 9228445 | Chr2 | 25382607 | LOC_Os02g42200 |
