Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1292-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1292-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 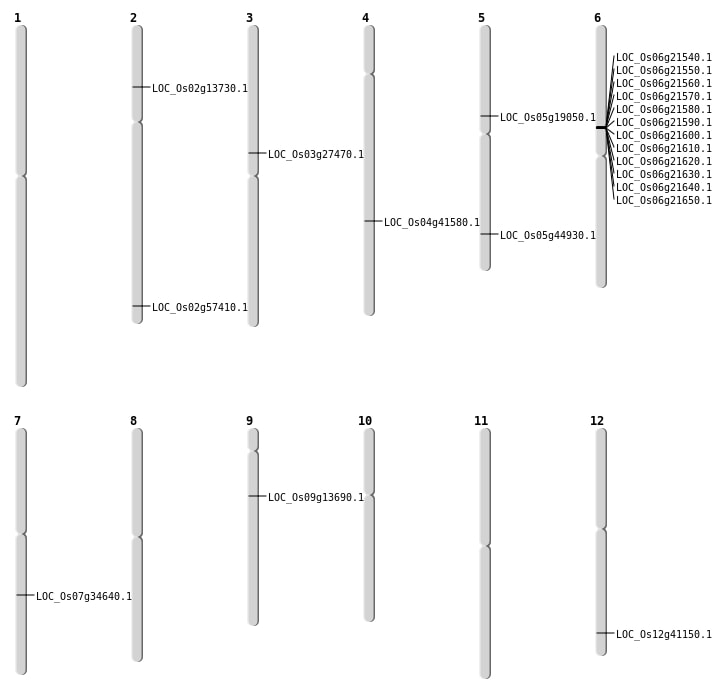
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14213141 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 4528804 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 4856909 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 5146685 | T-C | HET | INTRON | |
| SBS | Chr10 | 9598097 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25057786 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 23792927 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 17299069 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 21434083 | T-C | HET | INTRON | |
| SBS | Chr2 | 33756061 | G-A | HET | INTRON | |
| SBS | Chr2 | 4760785 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 7389082 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g13730 |
| SBS | Chr2 | 8406618 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 11506512 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 16141215 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 20965897 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 20965898 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 26055888 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16609268 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 30285354 | T-A | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr4 | 3288061 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 10424378 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 11056362 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19050 |
| SBS | Chr5 | 12594057 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26117153 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44930 |
| SBS | Chr5 | 9191649 | C-T | HET | INTRON | |
| SBS | Chr6 | 8182240 | C-A | HET | INTRON | |
| SBS | Chr7 | 20767441 | T-C | HET | SPLICE_SITE_DONOR | LOC_Os07g34640 |
| SBS | Chr8 | 10208467 | A-C | HOMO | INTRON | |
| SBS | Chr8 | 17547538 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 2321852 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 25152977 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 25152979 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 5506226 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 8007280 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g13690 |
| SBS | Chr9 | 8007283 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g13690 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2025734 | 2025746 | 12 | |
| Deletion | Chr1 | 5604842 | 5604843 | 1 | |
| Deletion | Chr11 | 5780756 | 5780765 | 9 | |
| Deletion | Chr6 | 8195327 | 8195328 | 1 | |
| Deletion | Chr12 | 11301115 | 11301116 | 1 | |
| Deletion | Chr6 | 12331986 | 12331987 | 1 | |
| Deletion | Chr6 | 12427001 | 12499000 | 72000 | 12 |
| Deletion | Chr5 | 13291047 | 13291062 | 15 | |
| Deletion | Chr3 | 15758705 | 15758706 | 1 | LOC_Os03g27470 |
| Deletion | Chr10 | 19302631 | 19302635 | 4 | |
| Deletion | Chr3 | 19663512 | 19663518 | 6 | |
| Deletion | Chr1 | 19805984 | 19805993 | 9 | |
| Deletion | Chr7 | 20232666 | 20232668 | 2 | |
| Deletion | Chr4 | 21199028 | 21199042 | 14 | |
| Deletion | Chr10 | 21436822 | 21436833 | 11 | |
| Deletion | Chr5 | 21917808 | 21917810 | 2 | |
| Deletion | Chr4 | 26699581 | 26699584 | 3 | |
| Deletion | Chr7 | 26732311 | 26732322 | 11 | |
| Deletion | Chr4 | 32383751 | 32383756 | 5 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2743564 | 2743566 | 3 | |
| Insertion | Chr8 | 27181136 | 27181136 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 35178476 | 35456568 | LOC_Os02g57410 |
| Inversion | Chr2 | 35178505 | 35456799 | LOC_Os02g57410 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 3893911 | Chr4 | 24669379 | LOC_Os04g41580 |
| Translocation | Chr7 | 11140853 | Chr2 | 19691663 | |
| Translocation | Chr7 | 15550451 | Chr5 | 5643368 | |
| Translocation | Chr12 | 25499852 | Chr4 | 15519255 | LOC_Os12g41150 |
