Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1293-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1293-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 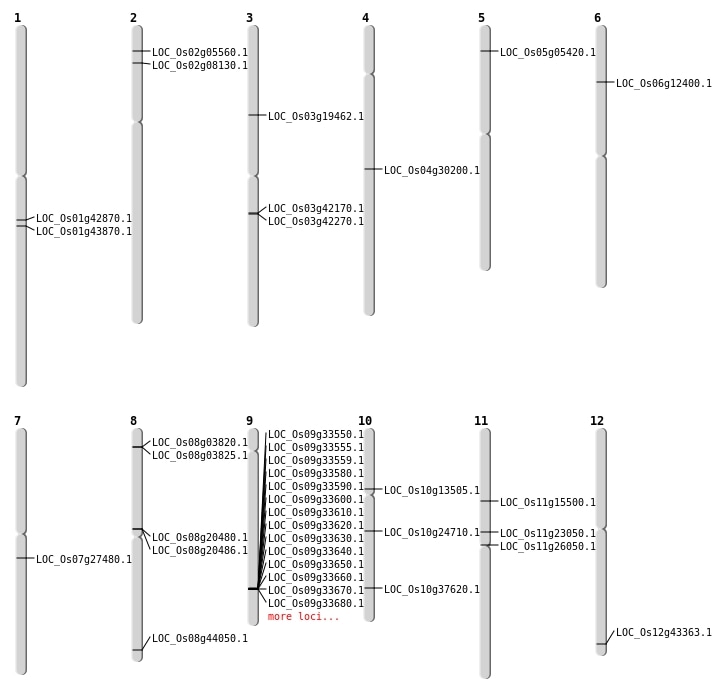
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13885855 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 37173534 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 12709322 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24710 |
| SBS | Chr11 | 13254405 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g23050 |
| SBS | Chr11 | 14903257 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26050 |
| SBS | Chr11 | 3822824 | G-A | HET | INTRON | |
| SBS | Chr12 | 26160870 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 26160871 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10167038 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 33411210 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 390124 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 4306382 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g08130 |
| SBS | Chr3 | 18197027 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 2281529 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 23461589 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g42170 |
| SBS | Chr3 | 23461590 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g42170 |
| SBS | Chr3 | 3030887 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 7983344 | T-A | HOMO | INTRON | |
| SBS | Chr4 | 23144813 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 8851877 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 2690596 | G-A | HET | STOP_GAINED | LOC_Os05g05420 |
| SBS | Chr5 | 4247129 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 5206565 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 14787929 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 22444668 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 9652891 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 23996410 | C-T | HOMO | UTR_3_PRIME |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 3647314 | 3647318 | 4 | |
| Deletion | Chr6 | 5549914 | 5549924 | 10 | |
| Deletion | Chr6 | 6734062 | 6734074 | 12 | LOC_Os06g12400 |
| Deletion | Chr10 | 7302551 | 7302561 | 10 | LOC_Os10g13505 |
| Deletion | Chr7 | 9535792 | 9535795 | 3 | |
| Deletion | Chr3 | 9992218 | 9992225 | 7 | |
| Deletion | Chr4 | 10811712 | 10811720 | 8 | |
| Deletion | Chr3 | 10945463 | 10945468 | 5 | LOC_Os03g19462 |
| Deletion | Chr7 | 11110255 | 11110419 | 164 | |
| Deletion | Chr8 | 12291001 | 12304000 | 13000 | 2 |
| Deletion | Chr8 | 12814905 | 12814931 | 26 | |
| Deletion | Chr1 | 13206730 | 13207032 | 302 | |
| Deletion | Chr5 | 15155064 | 15155066 | 2 | |
| Deletion | Chr7 | 15837631 | 15837641 | 10 | |
| Deletion | Chr4 | 17166187 | 17166201 | 14 | |
| Deletion | Chr9 | 19783001 | 19909000 | 126000 | 17 |
| Deletion | Chr2 | 22172988 | 22172989 | 1 | |
| Deletion | Chr3 | 23517237 | 23517249 | 12 | LOC_Os03g42270 |
| Deletion | Chr3 | 35009046 | 35009068 | 22 | |
| Deletion | Chr1 | 35475735 | 35475736 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 8782627 | 8782630 | 4 | LOC_Os11g15500 |
| Insertion | Chr2 | 17850754 | 17850755 | 2 | |
| Insertion | Chr6 | 7527996 | 7527996 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 15119388 | 16480442 | |
| Inversion | Chr7 | 15122871 | 16480430 | |
| Inversion | Chr10 | 20132060 | 20585022 | LOC_Os10g37620 |
| Inversion | Chr1 | 24395318 | 25135784 | 2 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1827307 | Chr7 | 10298338 | LOC_Os08g03820 |
| Translocation | Chr8 | 1828915 | Chr7 | 10298303 | LOC_Os08g03825 |
| Translocation | Chr11 | 7117859 | Chr4 | 18024546 | LOC_Os04g30200 |
| Translocation | Chr10 | 15476005 | Chr3 | 18904930 | |
| Translocation | Chr7 | 16017260 | Chr5 | 3266974 | LOC_Os07g27480 |
| Translocation | Chr12 | 26904451 | Chr1 | 13207027 | LOC_Os12g43363 |
| Translocation | Chr12 | 26904453 | Chr1 | 13206742 | LOC_Os12g43363 |
| Translocation | Chr8 | 27733813 | Chr2 | 2696906 | 2 |
| Translocation | Chr8 | 27741540 | Chr2 | 2696904 | LOC_Os02g05560 |
