Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1297-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1297-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 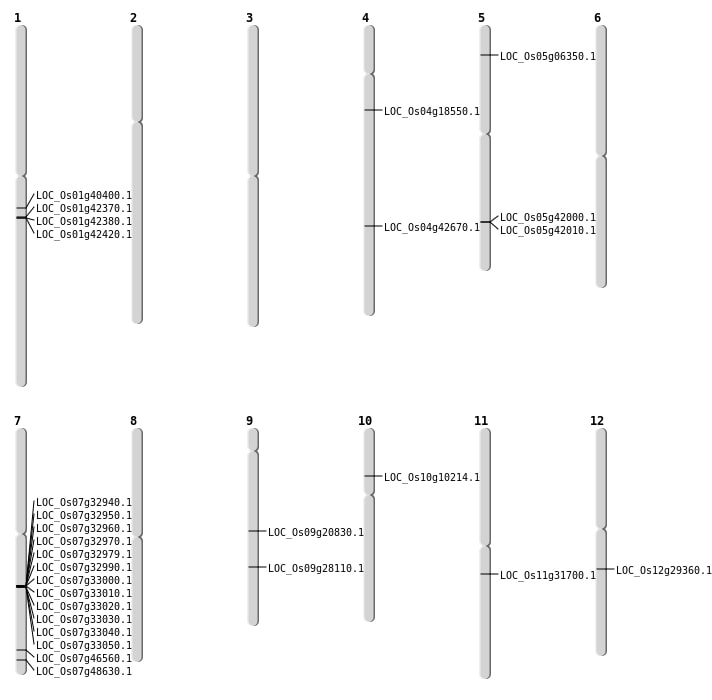
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15379999 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 16428825 | C-T | HET | INTRON | |
| SBS | Chr1 | 22092088 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22092089 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22330641 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 22808716 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g40400 |
| SBS | Chr1 | 25937524 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 41326493 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 41326496 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 11843143 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17369703 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 5213009 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 5619500 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g10214 |
| SBS | Chr11 | 18838096 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 21125400 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 4013338 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 10034894 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11450248 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 17431472 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29360 |
| SBS | Chr12 | 22401418 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 24242495 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 2703069 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 18435547 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 20424173 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 8939059 | G-C | HET | INTRON | |
| SBS | Chr3 | 11970420 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 18158365 | T-C | HET | INTRON | |
| SBS | Chr3 | 19983737 | C-T | HET | INTRON | |
| SBS | Chr3 | 6339645 | G-A | HET | INTRON | |
| SBS | Chr3 | 9863238 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 11420221 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 4007977 | C-T | HET | INTRON | |
| SBS | Chr5 | 29917910 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 13991390 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 15066808 | A-G | HET | INTRON | |
| SBS | Chr6 | 295981 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 30837448 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 1065828 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 2059855 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 22041444 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 27809377 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g46560 |
| SBS | Chr8 | 22283208 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 2241528 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 12081052 | T-C | HET | INTRON | |
| SBS | Chr9 | 12547825 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g20830 |
| SBS | Chr9 | 12999644 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 20428183 | T-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 20428184 | G-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 343189 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 5177567 | G-A | HET | INTRON |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 185752 | 185754 | 2 | |
| Deletion | Chr5 | 3252334 | 3252336 | 2 | LOC_Os05g06350 |
| Deletion | Chr10 | 3732004 | 3732019 | 15 | |
| Deletion | Chr11 | 3732560 | 3732562 | 2 | |
| Deletion | Chr9 | 4111973 | 4111975 | 2 | |
| Deletion | Chr12 | 7813860 | 7813861 | 1 | |
| Deletion | Chr1 | 8671975 | 8671986 | 11 | |
| Deletion | Chr1 | 8671981 | 8671992 | 11 | |
| Deletion | Chr10 | 9135070 | 9135071 | 1 | |
| Deletion | Chr12 | 15312238 | 15312242 | 4 | |
| Deletion | Chr3 | 16130547 | 16130555 | 8 | |
| Deletion | Chr9 | 17064804 | 17064816 | 12 | LOC_Os09g28110 |
| Deletion | Chr11 | 18556275 | 18556276 | 1 | LOC_Os11g31700 |
| Deletion | Chr10 | 19647742 | 19647749 | 7 | |
| Deletion | Chr7 | 19687001 | 19742000 | 55000 | 12 |
| Deletion | Chr5 | 19833130 | 19833134 | 4 | |
| Deletion | Chr10 | 20066996 | 20067015 | 19 | |
| Deletion | Chr2 | 20190500 | 20190501 | 1 | |
| Deletion | Chr3 | 20443536 | 20443538 | 2 | |
| Deletion | Chr10 | 21915056 | 21915057 | 1 | |
| Deletion | Chr1 | 22089100 | 22089104 | 4 | |
| Deletion | Chr1 | 24066349 | 24097402 | 31053 | 2 |
| Deletion | Chr1 | 24123454 | 24123465 | 11 | LOC_Os01g42420 |
| Deletion | Chr5 | 24589457 | 24589474 | 17 | 2 |
| Deletion | Chr4 | 25244934 | 25244935 | 1 | LOC_Os04g42670 |
| Deletion | Chr5 | 27914980 | 27914981 | 1 | |
| Deletion | Chr5 | 29074293 | 29074294 | 1 | |
| Deletion | Chr1 | 30451630 | 30451631 | 1 | |
| Deletion | Chr1 | 31231298 | 31231304 | 6 | |
| Deletion | Chr3 | 33955014 | 33955027 | 13 | |
| Deletion | Chr4 | 33980551 | 33980552 | 1 |
Insertions: 9
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 10248185 | 10528397 | LOC_Os04g18550 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 8980171 | Chr7 | 29117667 | LOC_Os07g48630 |
