Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1301-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1301-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 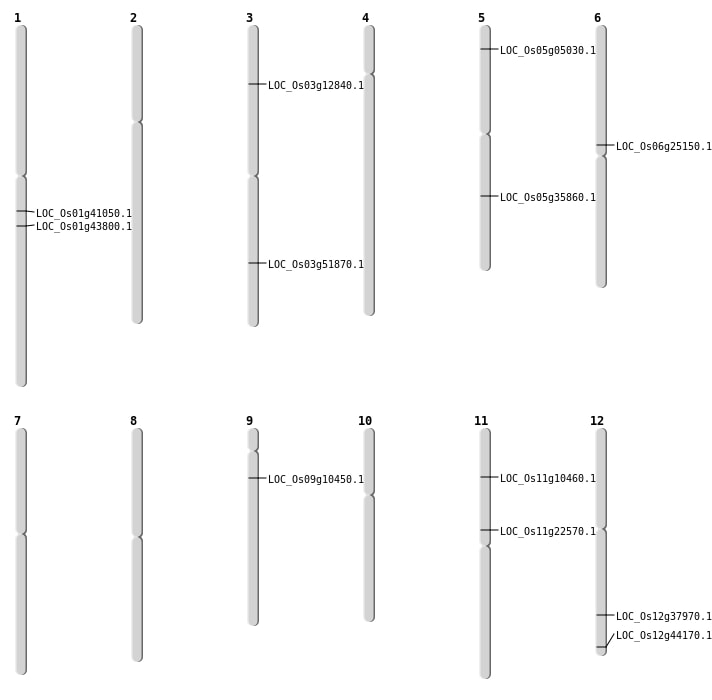
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17359372 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 25104552 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g43800 |
| SBS | Chr1 | 31884750 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 32952566 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 3763965 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 38253639 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 38450785 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 12982708 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22570 |
| SBS | Chr12 | 12015318 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 19361709 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 21045410 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 23385770 | T-A | HET | INTRON | |
| SBS | Chr12 | 23837333 | T-C | HET | INTRON | |
| SBS | Chr2 | 20586578 | G-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 35073305 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 22533267 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 29742500 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g51870 |
| SBS | Chr3 | 34998342 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 6902277 | T-C | HET | START_GAINED | LOC_Os03g12840 |
| SBS | Chr4 | 33503131 | G-A | HET | INTRON | |
| SBS | Chr4 | 7382806 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 11343911 | G-A | HET | INTRON | |
| SBS | Chr5 | 12270422 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 2449685 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g05030 |
| SBS | Chr5 | 7495522 | A-G | HET | INTRON | |
| SBS | Chr6 | 10133888 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 12034450 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 16678869 | C-T | HET | INTRON | |
| SBS | Chr6 | 27331790 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 28573325 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 5907655 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 23456894 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 27603968 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr9 | 9345783 | A-G | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 354361 | 354367 | 6 | |
| Deletion | Chr1 | 3763985 | 3763986 | 1 | |
| Deletion | Chr11 | 5700880 | 5700884 | 4 | LOC_Os11g10460 |
| Deletion | Chr9 | 5710657 | 5710662 | 5 | LOC_Os09g10450 |
| Deletion | Chr6 | 7109204 | 7109205 | 1 | |
| Deletion | Chr5 | 7202177 | 7202181 | 4 | |
| Deletion | Chr11 | 7213449 | 7213466 | 17 | |
| Deletion | Chr12 | 8660734 | 8660736 | 2 | |
| Deletion | Chr11 | 9919941 | 9919943 | 2 | |
| Deletion | Chr5 | 10839999 | 10840005 | 6 | |
| Deletion | Chr4 | 11793917 | 11793919 | 2 | |
| Deletion | Chr5 | 14149184 | 14149186 | 2 | |
| Deletion | Chr6 | 14713062 | 14714208 | 1146 | LOC_Os06g25150 |
| Deletion | Chr6 | 15256160 | 15256178 | 18 | |
| Deletion | Chr11 | 17370157 | 17370166 | 9 | |
| Deletion | Chr1 | 18025671 | 18025674 | 3 | |
| Deletion | Chr5 | 19287491 | 19287492 | 1 | |
| Deletion | Chr2 | 20192293 | 20192312 | 19 | |
| Deletion | Chr9 | 21041900 | 21041901 | 1 | |
| Deletion | Chr10 | 21240140 | 21240161 | 21 | |
| Deletion | Chr12 | 23326373 | 23326384 | 11 | LOC_Os12g37970 |
| Deletion | Chr8 | 26214860 | 26214861 | 1 | |
| Deletion | Chr12 | 27234724 | 27234728 | 4 | |
| Deletion | Chr8 | 27430237 | 27430252 | 15 | |
| Deletion | Chr3 | 34029957 | 34029959 | 2 | |
| Deletion | Chr2 | 35335537 | 35335541 | 4 | |
| Deletion | Chr3 | 36242148 | 36242166 | 18 | |
| Deletion | Chr1 | 36785108 | 36785116 | 8 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 200406 | 516186 | |
| Inversion | Chr5 | 21273209 | 21672082 | LOC_Os05g35860 |
| Inversion | Chr1 | 23208701 | 23247399 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 27401541 | Chr7 | 15223 | LOC_Os12g44170 |
