Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1302-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1302-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 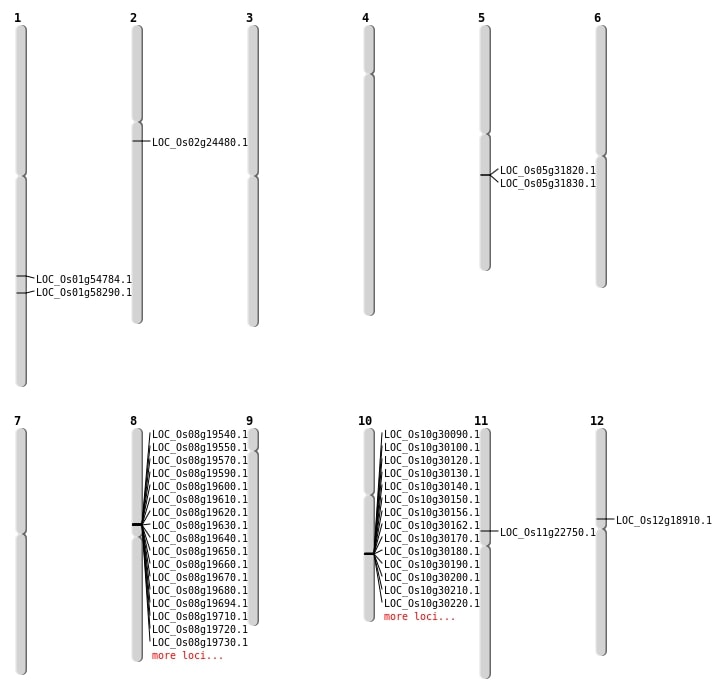
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12675262 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 36876593 | C-A | HET | INTRON | |
| SBS | Chr1 | 36876594 | G-A | HET | INTRON | |
| SBS | Chr1 | 38245212 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 40622375 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 40622376 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 42467522 | T-A | HET | INTRON | |
| SBS | Chr10 | 20037271 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 5180569 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 12117939 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 20214847 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 25548546 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 28869484 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 4847798 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 10975700 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g18910 |
| SBS | Chr12 | 11172212 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 26960265 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 7498991 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 14180410 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24480 |
| SBS | Chr2 | 17777627 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 21908884 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 29899485 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr3 | 14267113 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 31386319 | C-G | HET | INTRON | |
| SBS | Chr3 | 32615958 | A-G | HET | INTRON | |
| SBS | Chr4 | 12739586 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 17055212 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 19453073 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27758729 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 31586250 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 8831419 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 15037587 | C-T | HET | INTRON | |
| SBS | Chr6 | 15416478 | A-T | HET | INTRON | |
| SBS | Chr6 | 16886542 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 9145582 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19650634 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 4329907 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 7861992 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 10840283 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 11854817 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 11854819 | G-A | HOMO | INTRON |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 87834 | 87843 | 9 | |
| Deletion | Chr2 | 616225 | 616226 | 1 | |
| Deletion | Chr12 | 3708152 | 3708156 | 4 | |
| Deletion | Chr8 | 4116792 | 4116793 | 1 | |
| Deletion | Chr2 | 7003428 | 7003438 | 10 | |
| Deletion | Chr7 | 7703086 | 7703092 | 6 | |
| Deletion | Chr1 | 9005356 | 9005357 | 1 | |
| Deletion | Chr6 | 9144161 | 9144167 | 6 | |
| Deletion | Chr9 | 11046285 | 11046288 | 3 | |
| Deletion | Chr8 | 11683001 | 11848000 | 165000 | 21 |
| Deletion | Chr11 | 13085555 | 13085594 | 39 | LOC_Os11g22750 |
| Deletion | Chr12 | 14227011 | 14227027 | 16 | |
| Deletion | Chr9 | 14640821 | 14640827 | 6 | |
| Deletion | Chr5 | 14666715 | 14666717 | 2 | |
| Deletion | Chr8 | 15478920 | 15478932 | 12 | |
| Deletion | Chr10 | 15634001 | 15913000 | 279000 | 46 |
| Deletion | Chr3 | 16287372 | 16287376 | 4 | |
| Deletion | Chr7 | 16432322 | 16432323 | 1 | |
| Deletion | Chr4 | 16780925 | 16780926 | 1 | |
| Deletion | Chr5 | 18532771 | 18535822 | 3051 | 2 |
| Deletion | Chr1 | 21657453 | 21657455 | 2 | |
| Deletion | Chr4 | 22002254 | 22002260 | 6 | |
| Deletion | Chr5 | 26329720 | 26329721 | 1 | |
| Deletion | Chr1 | 29548537 | 29548568 | 31 | |
| Deletion | Chr1 | 31519143 | 31519153 | 10 | LOC_Os01g54784 |
| Deletion | Chr3 | 33650549 | 33650554 | 5 | |
| Deletion | Chr3 | 33655550 | 33655564 | 14 | |
| Deletion | Chr1 | 33692266 | 33692270 | 4 | LOC_Os01g58290 |
| Deletion | Chr1 | 33849806 | 33849808 | 2 | |
| Deletion | Chr4 | 35475450 | 35475454 | 4 | |
| Deletion | Chr1 | 36965173 | 36965174 | 1 |
Insertions: 5
No Inversion
No Translocation
