Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1304-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1304-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 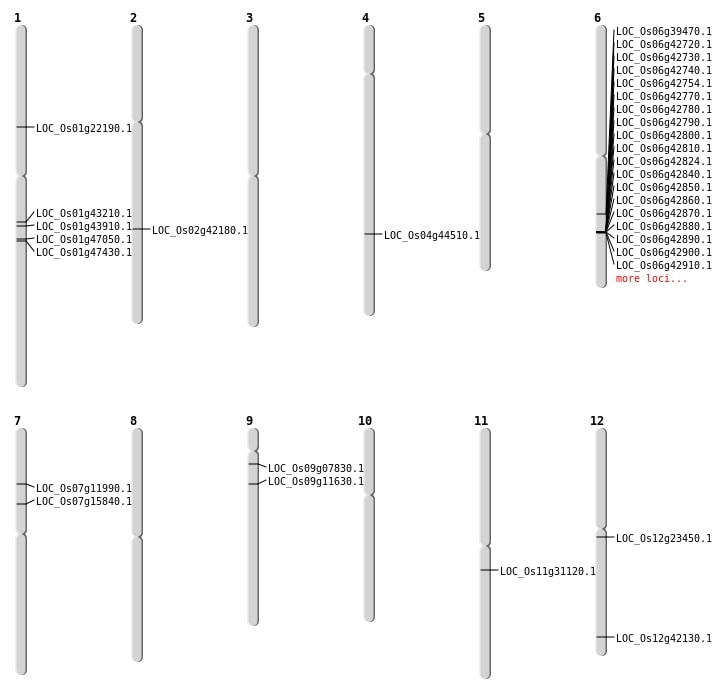
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12245106 | A-G | HET | INTRON | |
| SBS | Chr1 | 12475963 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g22190 |
| SBS | Chr1 | 14370429 | T-C | HET | INTRON | |
| SBS | Chr1 | 22511530 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 25079687 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 18694206 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7369280 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 18115342 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g31120 |
| SBS | Chr11 | 23049723 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 7598626 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 21543613 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 25138079 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26044535 | G-A | HET | INTRON | |
| SBS | Chr12 | 26121959 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42130 |
| SBS | Chr12 | 3538914 | C-T | HET | INTRON | |
| SBS | Chr2 | 10349583 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10349584 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14315557 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 32563866 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 33413874 | G-T | HET | INTRON | |
| SBS | Chr2 | 33674046 | T-C | HET | INTRON | |
| SBS | Chr3 | 15609969 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 16420228 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 19683147 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 19683148 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 25138720 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 25804780 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 12267199 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 17707437 | G-A | HET | INTRON | |
| SBS | Chr5 | 18853320 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 25080659 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 8084806 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 9329780 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 9922401 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 5768043 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 24226156 | G-A | HET | INTRON | |
| SBS | Chr7 | 6647608 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11990 |
| SBS | Chr7 | 9202531 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g15840 |
| SBS | Chr8 | 27943503 | A-T | HET | INTRON | |
| SBS | Chr9 | 19748393 | A-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 22053183 | G-A | HET | INTRON | |
| SBS | Chr9 | 3974206 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07830 |
| SBS | Chr9 | 5618349 | A-G | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 4762314 | 4762315 | 1 | |
| Deletion | Chr8 | 6583162 | 6583172 | 10 | |
| Deletion | Chr8 | 8232264 | 8232265 | 1 | |
| Deletion | Chr11 | 8850708 | 8850719 | 11 | |
| Deletion | Chr6 | 9606881 | 9606890 | 9 | |
| Deletion | Chr12 | 11724728 | 11724737 | 9 | |
| Deletion | Chr4 | 12226281 | 12226286 | 5 | |
| Deletion | Chr10 | 13747941 | 13747942 | 1 | |
| Deletion | Chr11 | 14166906 | 14166914 | 8 | |
| Deletion | Chr2 | 14399506 | 14399507 | 1 | |
| Deletion | Chr3 | 20441487 | 20441489 | 2 | |
| Deletion | Chr4 | 21382192 | 21382198 | 6 | |
| Deletion | Chr4 | 21560652 | 21560654 | 2 | |
| Deletion | Chr9 | 22916077 | 22916080 | 3 | |
| Deletion | Chr2 | 23100932 | 23100937 | 5 | |
| Deletion | Chr2 | 25370263 | 25370266 | 3 | LOC_Os02g42180 |
| Deletion | Chr6 | 25693001 | 25820000 | 127000 | 25 |
| Deletion | Chr12 | 26318130 | 26318133 | 3 | |
| Deletion | Chr4 | 26348270 | 26348273 | 3 | LOC_Os04g44510 |
| Deletion | Chr2 | 26987050 | 26987052 | 2 | |
| Deletion | Chr6 | 28286280 | 28286289 | 9 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 6704332 | 6704335 | 4 | |
| Insertion | Chr3 | 15159939 | 15159942 | 4 | |
| Insertion | Chr5 | 21243538 | 21243538 | 1 | |
| Insertion | Chr6 | 23438886 | 23438897 | 12 | LOC_Os06g39470 |
| Insertion | Chr9 | 10403603 | 10403604 | 2 | |
| Insertion | Chr9 | 15198129 | 15198132 | 4 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 5593998 | 6491167 | LOC_Os09g11630 |
| Inversion | Chr4 | 8091166 | 8094176 | |
| Inversion | Chr1 | 26872414 | 27107453 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 13265772 | Chr1 | 25152179 | 2 |
| Translocation | Chr12 | 13265774 | Chr1 | 24684367 | 2 |
| Translocation | Chr11 | 16996107 | Chr4 | 3614474 |
