Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1309-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1309-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 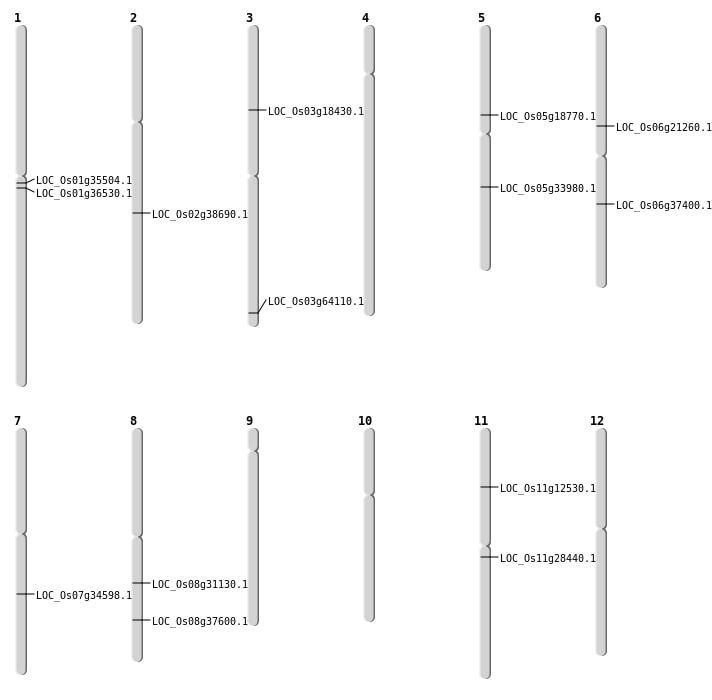
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10422438 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 19667889 | G-C | HOMO | INTRON | |
| SBS | Chr1 | 20276279 | T-A | HOMO | STOP_GAINED | LOC_Os01g36530 |
| SBS | Chr1 | 2549447 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 28967699 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1038640 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 16375193 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28440 |
| SBS | Chr11 | 18836550 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 3733354 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 7013142 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g12530 |
| SBS | Chr12 | 17329774 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 19421957 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 23390089 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g38690 |
| SBS | Chr2 | 4504653 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 5311527 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 6144154 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10339655 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 10339656 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 27775109 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 6314468 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 9389132 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 1032239 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 10345128 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1115679 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 14429626 | G-A | HET | INTRON | |
| SBS | Chr5 | 16891825 | A-T | HET | INTRON | |
| SBS | Chr5 | 7892642 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 22108473 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g37400 |
| SBS | Chr6 | 8748807 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12855067 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 5095591 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 19995242 | A-G | HET | INTRON | |
| SBS | Chr8 | 474147 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 19014733 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 3616042 | G-A | HOMO | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 6948413 | 6948426 | 13 | |
| Deletion | Chr5 | 7321116 | 7321129 | 13 | |
| Deletion | Chr3 | 10336214 | 10336257 | 43 | LOC_Os03g18430 |
| Deletion | Chr5 | 10885644 | 10885686 | 42 | LOC_Os05g18770 |
| Deletion | Chr4 | 11914447 | 11914448 | 1 | |
| Deletion | Chr3 | 12314931 | 12314934 | 3 | |
| Deletion | Chr6 | 12535460 | 12535461 | 1 | |
| Deletion | Chr4 | 13383336 | 13383340 | 4 | |
| Deletion | Chr7 | 13779058 | 13779069 | 11 | |
| Deletion | Chr1 | 13847138 | 13847139 | 1 | |
| Deletion | Chr6 | 16137363 | 16137374 | 11 | |
| Deletion | Chr8 | 17459178 | 17459196 | 18 | |
| Deletion | Chr8 | 19223228 | 19223234 | 6 | LOC_Os08g31130 |
| Deletion | Chr5 | 20062225 | 20062228 | 3 | LOC_Os05g33980 |
| Deletion | Chr3 | 20208150 | 20208159 | 9 | |
| Deletion | Chr4 | 20313319 | 20313322 | 3 | |
| Deletion | Chr8 | 23676640 | 23676644 | 4 | |
| Deletion | Chr6 | 24491076 | 24491079 | 3 | |
| Deletion | Chr1 | 25493635 | 25493636 | 1 | |
| Deletion | Chr4 | 25926429 | 25926441 | 12 | |
| Deletion | Chr3 | 33811482 | 33811485 | 3 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11052183 | 11052184 | 2 | |
| Insertion | Chr11 | 1878359 | 1878360 | 2 | |
| Insertion | Chr5 | 49552 | 49553 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 12292645 | 13288458 | LOC_Os06g21260 |
| Inversion | Chr6 | 12297726 | 13288459 | |
| Inversion | Chr7 | 15159329 | 15667779 | |
| Inversion | Chr1 | 19264221 | 19645709 | LOC_Os01g35504 |
| Inversion | Chr3 | 36140513 | 36229621 | LOC_Os03g64110 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 20745755 | Chr2 | 15495233 | LOC_Os07g34598 |
| Translocation | Chr8 | 23824300 | Chr7 | 20745764 | 2 |
| Translocation | Chr11 | 26515493 | Chr8 | 23824337 | LOC_Os08g37600 |
| Translocation | Chr11 | 26517206 | Chr8 | 23824961 | LOC_Os08g37600 |
