Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1320-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1320-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 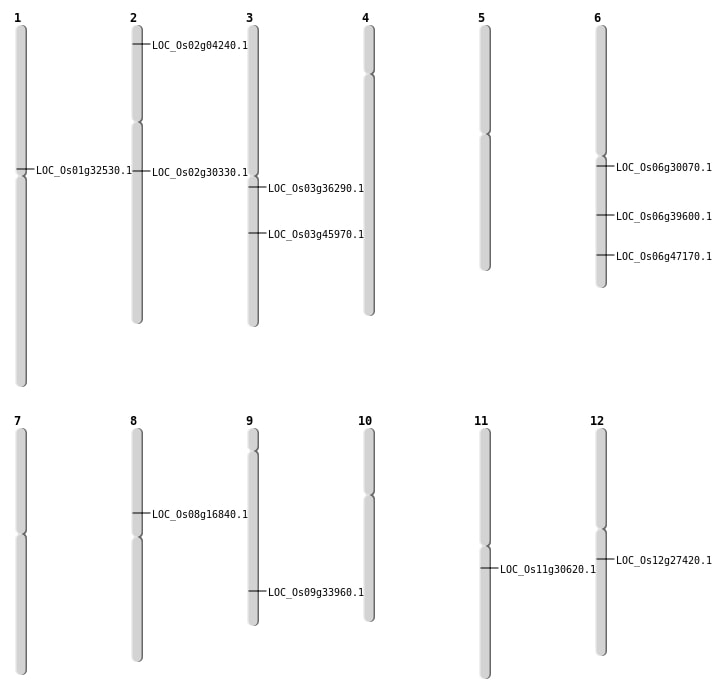
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31950482 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 32784414 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 37080190 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 37080191 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11487417 | C-T | HET | INTRON | |
| SBS | Chr10 | 16054844 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 1651793 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 16607758 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 17566145 | C-T | HOMO | UTR_5_PRIME | |
| SBS | Chr10 | 18768490 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 2531079 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 16828360 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 22749059 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 15313220 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 1189726 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 20129686 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g36290 |
| SBS | Chr3 | 24739053 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 34352986 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3903367 | G-T | HET | INTRON | |
| SBS | Chr3 | 8452349 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 11647579 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 13056714 | C-T | HET | INTRON | |
| SBS | Chr4 | 19559000 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 17013722 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 25857621 | A-T | HET | INTRON | |
| SBS | Chr6 | 12672722 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17328008 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g30070 |
| SBS | Chr6 | 17652576 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 28604747 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr6 | 29420836 | C-G | HOMO | INTRON | |
| SBS | Chr7 | 20789898 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr7 | 20789899 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr7 | 27683597 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 8437178 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 21585785 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 22202680 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 3193747 | G-A | HET | INTRON | |
| SBS | Chr9 | 13805119 | C-G | HOMO | INTRON |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 260909 | 260911 | 2 | |
| Deletion | Chr10 | 448423 | 448429 | 6 | |
| Deletion | Chr2 | 1863267 | 1863275 | 8 | LOC_Os02g04240 |
| Deletion | Chr10 | 3420841 | 3420848 | 7 | |
| Deletion | Chr2 | 4584707 | 4584708 | 1 | |
| Deletion | Chr2 | 9136631 | 9136741 | 110 | |
| Deletion | Chr8 | 10296877 | 10296880 | 3 | LOC_Os08g16840 |
| Deletion | Chr7 | 10319251 | 10319258 | 7 | |
| Deletion | Chr6 | 12394104 | 12394227 | 123 | |
| Deletion | Chr12 | 16139898 | 16139913 | 15 | LOC_Os12g27420 |
| Deletion | Chr5 | 16409569 | 16409593 | 24 | |
| Deletion | Chr1 | 17843002 | 17843008 | 6 | LOC_Os01g32530 |
| Deletion | Chr2 | 18069617 | 18069665 | 48 | LOC_Os02g30330 |
| Deletion | Chr9 | 20046395 | 20046400 | 5 | LOC_Os09g33960 |
| Deletion | Chr5 | 23195947 | 23195955 | 8 | |
| Deletion | Chr7 | 26107017 | 26107019 | 2 | |
| Deletion | Chr5 | 28297593 | 28297594 | 1 | |
| Deletion | Chr4 | 33817768 | 33817784 | 16 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr6 | 5232124 | 5232124 | 1 | |
| Insertion | Chr7 | 4780597 | 4780598 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 17830017 | 17834052 | LOC_Os11g30620 |
| Inversion | Chr6 | 23505507 | 23506059 | LOC_Os06g39600 |
| Inversion | Chr1 | 37159624 | 37162967 | |
| Inversion | Chr1 | 37159635 | 37162971 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 16156152 | Chr3 | 25977074 | |
| Translocation | Chr8 | 16156163 | Chr3 | 25988662 | LOC_Os03g45970 |
