Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1322-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1322-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 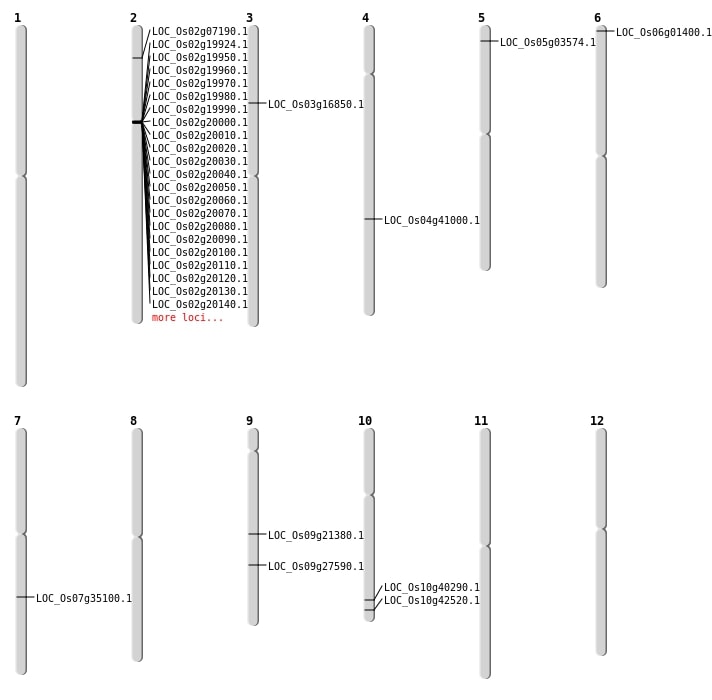
|
| Variant Information |
|---|
Single base substitutions: 21
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19949028 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 12407415 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 11632792 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 20404988 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 6353552 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 19894239 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 20878486 | A-T | HET | INTRON | |
| SBS | Chr2 | 2123808 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 9502480 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 19357729 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29450160 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr5 | 10037615 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 22039154 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 241828 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g01400 |
| SBS | Chr7 | 17680150 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 21026407 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g35100 |
| SBS | Chr8 | 27181345 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 10522923 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr9 | 12670968 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 12699826 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 15452477 | A-T | HET | INTRON |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 978590 | 978614 | 24 | |
| Deletion | Chr5 | 1517809 | 1517815 | 6 | LOC_Os05g03574 |
| Deletion | Chr10 | 2233698 | 2233730 | 32 | |
| Deletion | Chr12 | 3134922 | 3134923 | 1 | |
| Deletion | Chr9 | 3590083 | 3590095 | 12 | |
| Deletion | Chr9 | 3635021 | 3635032 | 11 | |
| Deletion | Chr2 | 3697505 | 3697508 | 3 | LOC_Os02g07190 |
| Deletion | Chr3 | 9359541 | 9359544 | 3 | LOC_Os03g16850 |
| Deletion | Chr2 | 11715001 | 12292000 | 577000 | 83 |
| Deletion | Chr7 | 11897199 | 11897202 | 3 | |
| Deletion | Chr9 | 12914047 | 12914053 | 6 | LOC_Os09g21380 |
| Deletion | Chr9 | 16766173 | 16766177 | 4 | LOC_Os09g27590 |
| Deletion | Chr7 | 17459989 | 17459992 | 3 | |
| Deletion | Chr9 | 18746135 | 18746138 | 3 | |
| Deletion | Chr9 | 19373004 | 19373007 | 3 | |
| Deletion | Chr2 | 19510001 | 19590000 | 80000 | 11 |
| Deletion | Chr2 | 20824797 | 20824801 | 4 | LOC_Os02g34710 |
| Deletion | Chr10 | 21599073 | 21599084 | 11 | LOC_Os10g40290 |
| Deletion | Chr10 | 22933805 | 22937203 | 3398 | LOC_Os10g42520 |
| Deletion | Chr4 | 24331204 | 24331227 | 23 | LOC_Os04g41000 |
| Deletion | Chr8 | 27105496 | 27105499 | 3 |
No Insertion
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 11715248 | 12294294 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 5208106 | Chr2 | 12292266 |
