Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1327-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1327-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 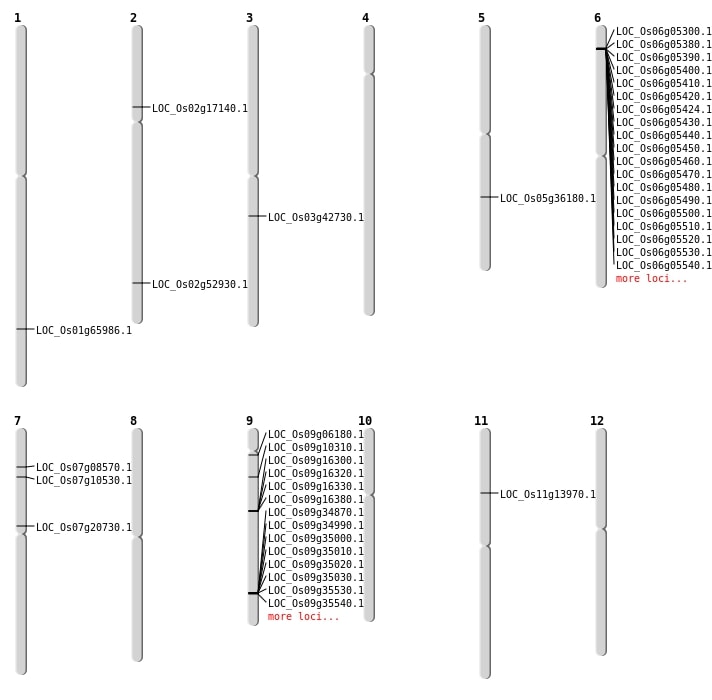
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 39049603 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 2039964 | A-C | HOMO | INTRON | |
| SBS | Chr11 | 14212929 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16112098 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 4906677 | A-G | HET | INTRON | |
| SBS | Chr12 | 8599564 | A-G | HET | INTRON | |
| SBS | Chr12 | 9699801 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 32359566 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g52930 |
| SBS | Chr2 | 5318590 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9820279 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g17140 |
| SBS | Chr4 | 10122523 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14564992 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21445796 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g36180 |
| SBS | Chr5 | 26805522 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 5163619 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 2386149 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g05300 |
| SBS | Chr6 | 30530507 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 19708422 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 4917037 | T-A | HET | INTRON | |
| SBS | Chr7 | 5700379 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12687840 | C-A | HET | INTRON | |
| SBS | Chr8 | 12687841 | A-T | HET | INTRON | |
| SBS | Chr8 | 18482938 | T-A | HET | INTRON | |
| SBS | Chr8 | 5537140 | G-A | HET | INTRON | |
| SBS | Chr9 | 10013582 | A-G | HET | INTRON | |
| SBS | Chr9 | 10019867 | C-T | HET | INTRON | |
| SBS | Chr9 | 10475684 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 10827665 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 1787702 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 18722414 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 5621156 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10310 |
| SBS | Chr9 | 6237020 | T-C | HET | INTRON |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1312726 | 1312727 | 1 | |
| Deletion | Chr6 | 2428001 | 2522000 | 94000 | 21 |
| Deletion | Chr9 | 2896763 | 2896771 | 8 | LOC_Os09g06180 |
| Deletion | Chr7 | 4885546 | 4885547 | 1 | |
| Deletion | Chr12 | 4950405 | 4950625 | 220 | |
| Deletion | Chr7 | 5674830 | 5674884 | 54 | |
| Deletion | Chr10 | 5733338 | 5733405 | 67 | |
| Deletion | Chr11 | 7746358 | 7746392 | 34 | LOC_Os11g13970 |
| Deletion | Chr1 | 8781943 | 8781947 | 4 | |
| Deletion | Chr9 | 9957811 | 9970810 | 12999 | 3 |
| Deletion | Chr9 | 10013499 | 10013522 | 23 | LOC_Os09g16380 |
| Deletion | Chr6 | 11319133 | 11319134 | 1 | |
| Deletion | Chr4 | 11552039 | 11552048 | 9 | |
| Deletion | Chr11 | 14295487 | 14295489 | 2 | |
| Deletion | Chr4 | 14632936 | 14632937 | 1 | |
| Deletion | Chr5 | 15021356 | 15021364 | 8 | |
| Deletion | Chr6 | 15720001 | 15791000 | 71000 | 11 |
| Deletion | Chr6 | 16638001 | 16690000 | 52000 | 9 |
| Deletion | Chr8 | 16639419 | 16639425 | 6 | |
| Deletion | Chr1 | 19129890 | 19129904 | 14 | |
| Deletion | Chr8 | 19696326 | 19696362 | 36 | |
| Deletion | Chr9 | 20323231 | 20323232 | 1 | LOC_Os09g34870 |
| Deletion | Chr9 | 20387275 | 20591362 | 204087 | 31 |
| Deletion | Chr8 | 20539291 | 20539299 | 8 | |
| Deletion | Chr5 | 23207468 | 23207470 | 2 | |
| Deletion | Chr3 | 25160343 | 25160351 | 8 | |
| Deletion | Chr11 | 25430310 | 25430325 | 15 | |
| Deletion | Chr3 | 27009891 | 27009892 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 32044669 | 32044669 | 1 | |
| Insertion | Chr1 | 38794092 | 38794092 | 1 | |
| Insertion | Chr6 | 8965726 | 8965726 | 1 | |
| Insertion | Chr9 | 20595580 | 20595582 | 3 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 2522156 | 3764709 | 2 |
| Inversion | Chr7 | 4425702 | 5673842 | LOC_Os07g08570 |
| Inversion | Chr7 | 4425717 | 5683075 | 2 |
| Inversion | Chr3 | 23789971 | 23886516 | LOC_Os03g42730 |
| Inversion | Chr3 | 23810276 | 23886505 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 5673858 | Chr1 | 38301839 | LOC_Os01g65986 |
| Translocation | Chr7 | 5683052 | Chr1 | 38301817 | 2 |
| Translocation | Chr7 | 11991485 | Chr6 | 15790652 | LOC_Os07g20730 |
