Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1330-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1330-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 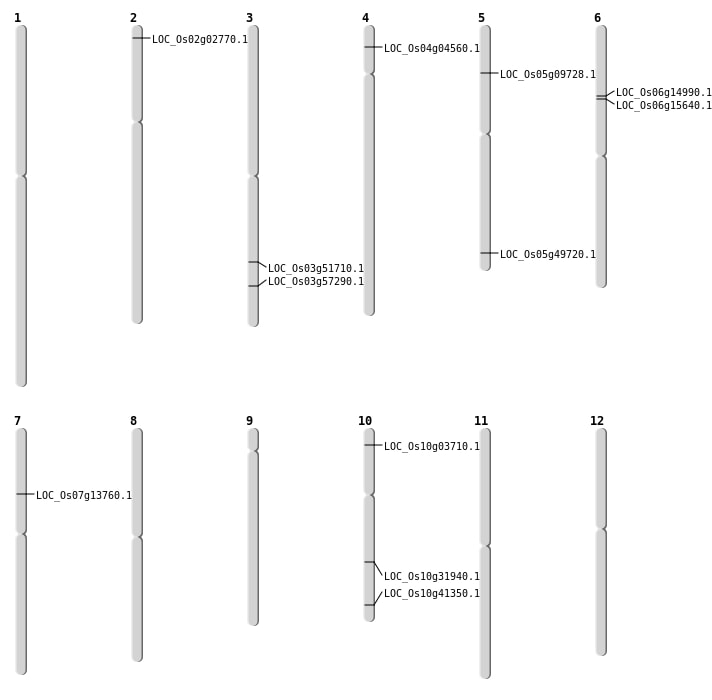
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28293640 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 37183701 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 2017811 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 22224807 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g41350 |
| SBS | Chr10 | 22785591 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7399513 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 13804634 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 14183016 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15410638 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 6040511 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 8982840 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 23779575 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 3010817 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 31969642 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 35338858 | T-C | HET | INTRON | |
| SBS | Chr2 | 6218892 | T-C | HET | INTRON | |
| SBS | Chr3 | 26361792 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 10584426 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 2180997 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04560 |
| SBS | Chr4 | 2263648 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 25499016 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 27939345 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr4 | 905883 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 1139609 | A-T | HOMO | INTRON | |
| SBS | Chr5 | 15072795 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 28456541 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 28737882 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr5 | 5512492 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09728 |
| SBS | Chr6 | 12200822 | A-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 20565593 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 26405399 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 26985313 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 27857180 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 29433147 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 30792285 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 4082097 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 10519098 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 18272485 | G-A | HET | INTRON | |
| SBS | Chr8 | 9429667 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 14725821 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 18359896 | G-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 986561 | 986564 | 3 | |
| Deletion | Chr2 | 1047681 | 1047743 | 62 | LOC_Os02g02770 |
| Deletion | Chr9 | 2329521 | 2329523 | 2 | |
| Deletion | Chr11 | 4138501 | 4138511 | 10 | |
| Deletion | Chr7 | 7896513 | 7896521 | 8 | LOC_Os07g13760 |
| Deletion | Chr11 | 8392836 | 8392837 | 1 | |
| Deletion | Chr6 | 8501935 | 8501948 | 13 | LOC_Os06g14990 |
| Deletion | Chr6 | 8857794 | 8860457 | 2663 | LOC_Os06g15640 |
| Deletion | Chr12 | 10132146 | 10132195 | 49 | |
| Deletion | Chr11 | 10246189 | 10246192 | 3 | |
| Deletion | Chr4 | 11639104 | 11639140 | 36 | |
| Deletion | Chr9 | 12012741 | 12012742 | 1 | |
| Deletion | Chr2 | 14051532 | 14051945 | 413 | |
| Deletion | Chr10 | 17712880 | 17712884 | 4 | |
| Deletion | Chr1 | 18798212 | 18798214 | 2 | |
| Deletion | Chr1 | 18798216 | 18798218 | 2 | |
| Deletion | Chr2 | 19161945 | 19161955 | 10 | |
| Deletion | Chr1 | 21331627 | 21331628 | 1 | |
| Deletion | Chr4 | 24662681 | 24662690 | 9 | |
| Deletion | Chr12 | 25714784 | 25714790 | 6 | |
| Deletion | Chr11 | 27151608 | 27151611 | 3 | |
| Deletion | Chr3 | 29631698 | 29631702 | 4 | LOC_Os03g51710 |
| Deletion | Chr1 | 31756442 | 31756459 | 17 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23798371 | 23798372 | 2 | |
| Insertion | Chr1 | 42960941 | 42960963 | 23 | |
| Insertion | Chr2 | 7763376 | 7763377 | 2 | |
| Insertion | Chr6 | 700093 | 700094 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 10042277 | 10774445 | |
| Inversion | Chr8 | 10059054 | 10774457 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1494099 | Chr3 | 32677483 | LOC_Os03g57290 |
| Translocation | Chr10 | 1674878 | Chr3 | 32677089 | 2 |
| Translocation | Chr12 | 26819090 | Chr10 | 16769335 | LOC_Os10g31940 |
| Translocation | Chr5 | 28512086 | Chr3 | 32677090 | 2 |
| Translocation | Chr5 | 28512102 | Chr3 | 32677514 | 2 |
