Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1337-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1337-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 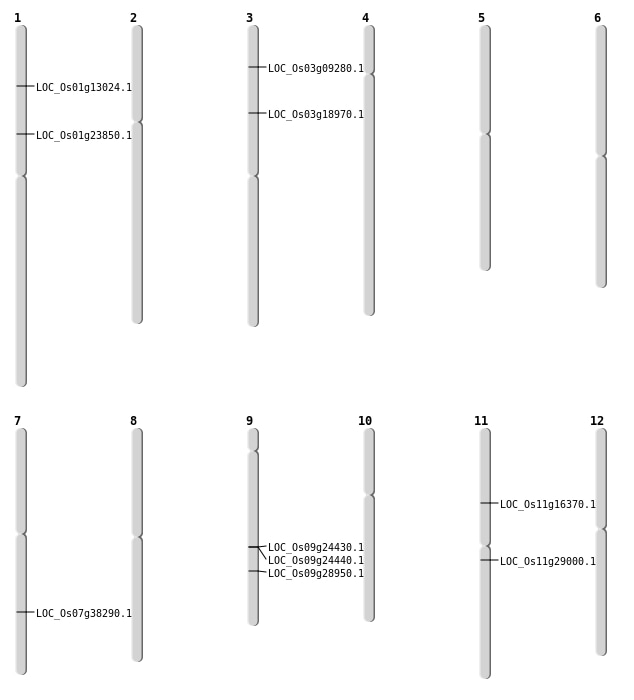
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13424444 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g23850 |
| SBS | Chr1 | 20699972 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 31819458 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 6263252 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 6804463 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 12376575 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 20948376 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 26644664 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 8195759 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 15655556 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 20073900 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 20707974 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 27725443 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 30581576 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 35746567 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 8241872 | A-G | HOMO | INTRON | |
| SBS | Chr3 | 16136843 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 18817260 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 2820941 | G-C | HOMO | INTRON | |
| SBS | Chr4 | 3142329 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 27250746 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20856018 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 3940326 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 8963493 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 8963494 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 11936937 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 13302681 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 29051731 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 1060299 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 16113494 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5194750 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 13296669 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 1375062 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 4481324 | A-T | HET | INTRON |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 146060 | 146097 | 37 | |
| Deletion | Chr10 | 1344059 | 1344078 | 19 | |
| Deletion | Chr6 | 1659705 | 1659713 | 8 | |
| Deletion | Chr9 | 1836377 | 1836378 | 1 | |
| Deletion | Chr4 | 4449928 | 4449931 | 3 | |
| Deletion | Chr3 | 4850778 | 4850781 | 3 | LOC_Os03g09280 |
| Deletion | Chr9 | 5084012 | 5084013 | 1 | |
| Deletion | Chr5 | 6313160 | 6313165 | 5 | |
| Deletion | Chr1 | 7245002 | 7245007 | 5 | LOC_Os01g13024 |
| Deletion | Chr12 | 8110932 | 8110938 | 6 | |
| Deletion | Chr11 | 8369869 | 8369871 | 2 | |
| Deletion | Chr11 | 9036421 | 9037518 | 1097 | LOC_Os11g16370 |
| Deletion | Chr9 | 9059114 | 9059132 | 18 | |
| Deletion | Chr3 | 10622085 | 10622086 | 1 | LOC_Os03g18970 |
| Deletion | Chr10 | 10698029 | 10698043 | 14 | |
| Deletion | Chr4 | 10740446 | 10740458 | 12 | |
| Deletion | Chr1 | 10976719 | 10976730 | 11 | |
| Deletion | Chr9 | 14527508 | 14541126 | 13618 | 2 |
| Deletion | Chr11 | 14992007 | 14992008 | 1 | |
| Deletion | Chr9 | 16740897 | 16740905 | 8 | |
| Deletion | Chr11 | 16781983 | 16781999 | 16 | LOC_Os11g29000 |
| Deletion | Chr3 | 20893927 | 20893929 | 2 | |
| Deletion | Chr2 | 20944258 | 20944268 | 10 | |
| Deletion | Chr6 | 21416308 | 21416309 | 1 | |
| Deletion | Chr3 | 22122650 | 22122651 | 1 | |
| Deletion | Chr7 | 22319895 | 22319897 | 2 | |
| Deletion | Chr3 | 24679919 | 24679939 | 20 | |
| Deletion | Chr3 | 24679921 | 24679941 | 20 | |
| Deletion | Chr2 | 29001806 | 29001817 | 11 | |
| Deletion | Chr1 | 29656421 | 29656426 | 5 | |
| Deletion | Chr1 | 31347551 | 31347554 | 3 | |
| Deletion | Chr3 | 32977514 | 32977515 | 1 | |
| Deletion | Chr4 | 34690308 | 34690423 | 115 | |
| Deletion | Chr2 | 35474721 | 35477400 | 2679 |
Insertions: 7
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 13545546 | 13551459 | |
| Inversion | Chr9 | 13545572 | 13551463 | |
| Inversion | Chr9 | 17343140 | 17614700 | LOC_Os09g28950 |
| Inversion | Chr7 | 19462158 | 23001384 | LOC_Os07g38290 |
| Inversion | Chr7 | 23001386 | 24032228 | LOC_Os07g38290 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 9036443 | Chr3 | 29223375 | LOC_Os11g16370 |
| Translocation | Chr11 | 9037519 | Chr3 | 29223369 | LOC_Os11g16370 |
| Translocation | Chr4 | 15896402 | Chr1 | 9105546 | |
| Translocation | Chr9 | 16416198 | Chr2 | 2673915 |
