Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1344-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1344-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 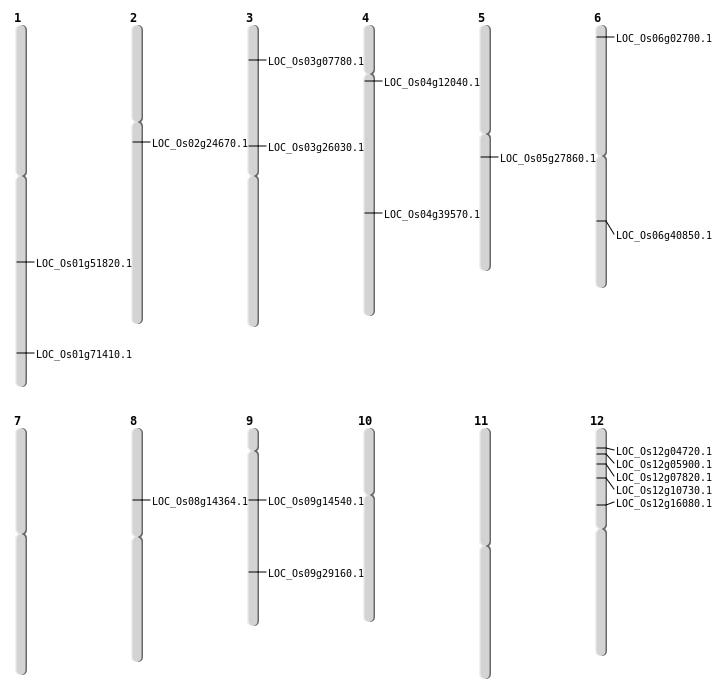
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13893440 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 14094773 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 14094774 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 20592598 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 28962461 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15130594 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 4829318 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 13497858 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 15332902 | C-T | HET | INTRON | |
| SBS | Chr12 | 2776623 | T-C | HET | INTRON | |
| SBS | Chr12 | 5761717 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g10730 |
| SBS | Chr2 | 14310782 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24670 |
| SBS | Chr2 | 33542731 | A-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr3 | 17169068 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 18699629 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 20455478 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 3953204 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g07780 |
| SBS | Chr3 | 7630567 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14547660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27070072 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 6607918 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12040 |
| SBS | Chr5 | 16243362 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g27860 |
| SBS | Chr5 | 3579783 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 8882143 | G-T | HET | INTRON | |
| SBS | Chr6 | 4417217 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 9190776 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19681313 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 7654026 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 27749671 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 18938461 | T-A | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr9 | 2898033 | G-C | HET | INTRON | |
| SBS | Chr9 | 8604140 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14540 |
| SBS | Chr9 | 8604141 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14540 |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 287168 | 287169 | 1 | |
| Deletion | Chr6 | 942675 | 942686 | 11 | |
| Deletion | Chr6 | 967437 | 967444 | 7 | LOC_Os06g02700 |
| Deletion | Chr12 | 2828310 | 2828318 | 8 | |
| Deletion | Chr2 | 4526405 | 4526421 | 16 | |
| Deletion | Chr10 | 5962608 | 5963169 | 561 | |
| Deletion | Chr1 | 7374745 | 7374993 | 248 | |
| Deletion | Chr2 | 8038438 | 8038439 | 1 | |
| Deletion | Chr2 | 8280381 | 8280387 | 6 | |
| Deletion | Chr8 | 8617491 | 8617501 | 10 | LOC_Os08g14364 |
| Deletion | Chr12 | 9179792 | 9179793 | 1 | LOC_Os12g16080 |
| Deletion | Chr12 | 9501178 | 9501181 | 3 | |
| Deletion | Chr3 | 10970442 | 10970443 | 1 | |
| Deletion | Chr12 | 11826805 | 11826852 | 47 | |
| Deletion | Chr12 | 13349197 | 13349216 | 19 | |
| Deletion | Chr3 | 13413549 | 13413550 | 1 | |
| Deletion | Chr3 | 14922307 | 14922308 | 1 | LOC_Os03g26030 |
| Deletion | Chr9 | 17732420 | 17732422 | 2 | LOC_Os09g29160 |
| Deletion | Chr6 | 20819562 | 20819566 | 4 | |
| Deletion | Chr9 | 21184594 | 21184600 | 6 | |
| Deletion | Chr5 | 22163630 | 22163631 | 1 | |
| Deletion | Chr3 | 22556378 | 22556379 | 1 | |
| Deletion | Chr4 | 23579832 | 23579838 | 6 | LOC_Os04g39570 |
| Deletion | Chr4 | 26927639 | 26930380 | 2741 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr4 | 35096487 | 35096508 | 21 | |
| Deletion | Chr1 | 41375337 | 41375341 | 4 | LOC_Os01g71410 |
Insertions: 6
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 2010399 | 3957605 | 2 |
| Inversion | Chr12 | 2711895 | 2715133 | LOC_Os12g05900 |
| Inversion | Chr12 | 5517220 | 5765055 | |
| Inversion | Chr1 | 29794961 | 37075544 | LOC_Os01g51820 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2127588 | Chr1 | 37075557 | |
| Translocation | Chr11 | 2127760 | Chr1 | 29794970 | LOC_Os01g51820 |
| Translocation | Chr12 | 5765054 | Chr6 | 29546362 | |
| Translocation | Chr12 | 5765327 | Chr6 | 29546356 | |
| Translocation | Chr10 | 5962610 | Chr3 | 25741964 | |
| Translocation | Chr10 | 5963212 | Chr3 | 25742005 | |
| Translocation | Chr8 | 27702792 | Chr6 | 24365551 | LOC_Os06g40850 |
