Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1345-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1345-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 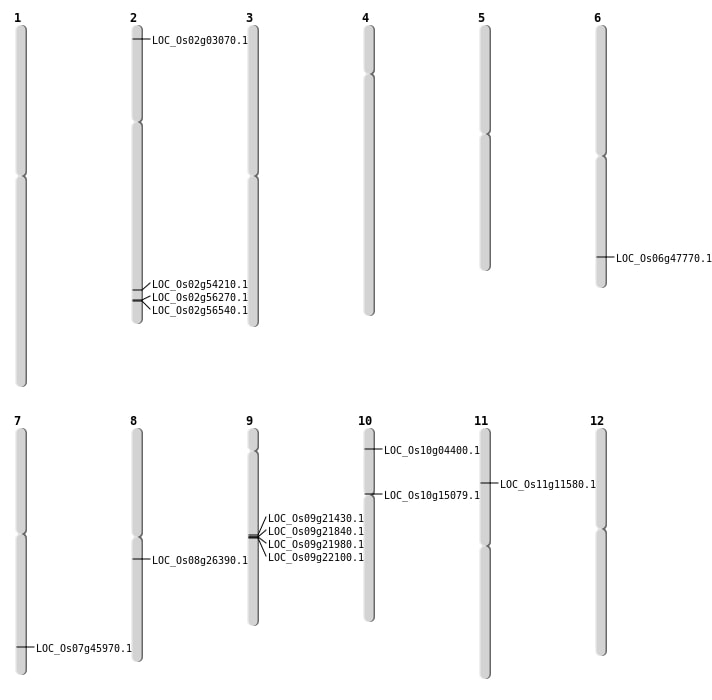
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 6882925 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 7895494 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g15079 |
| SBS | Chr11 | 6413329 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 15613360 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 23144572 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 23186257 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 28040208 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 33236351 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g54210 |
| SBS | Chr2 | 34429148 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56270 |
| SBS | Chr2 | 34633788 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56540 |
| SBS | Chr2 | 8624779 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 8678152 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 9111796 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 19075191 | C-A | HET | INTRON | |
| SBS | Chr3 | 19075192 | T-A | HET | INTRON | |
| SBS | Chr3 | 32333342 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 35753690 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 17667347 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 19494658 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 21545811 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 31710289 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 17131250 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 19590963 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 12858137 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 28922639 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 16082487 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g26390 |
| SBS | Chr8 | 17405004 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 23546327 | T-G | HET | INTRON | |
| SBS | Chr8 | 24053416 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 25612133 | A-C | HET | INTRON | |
| SBS | Chr8 | 25612134 | G-C | HET | INTRON | |
| SBS | Chr8 | 8563740 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19759792 | G-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 20826765 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 9586608 | C-T | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 427186 | 427198 | 12 | |
| Deletion | Chr2 | 1217830 | 1217842 | 12 | LOC_Os02g03070 |
| Deletion | Chr10 | 2233534 | 2233565 | 31 | |
| Deletion | Chr8 | 2242503 | 2242514 | 11 | |
| Deletion | Chr1 | 3526557 | 3526564 | 7 | |
| Deletion | Chr11 | 6357276 | 6357315 | 39 | |
| Deletion | Chr11 | 6449315 | 6449327 | 12 | LOC_Os11g11580 |
| Deletion | Chr2 | 8203583 | 8203596 | 13 | |
| Deletion | Chr8 | 8334043 | 8334045 | 2 | |
| Deletion | Chr11 | 8616174 | 8616176 | 2 | |
| Deletion | Chr3 | 8812919 | 8812925 | 6 | |
| Deletion | Chr2 | 10888892 | 10888909 | 17 | |
| Deletion | Chr9 | 14964193 | 14964222 | 29 | |
| Deletion | Chr2 | 18547807 | 18547808 | 1 | |
| Deletion | Chr1 | 18620683 | 18620692 | 9 | |
| Deletion | Chr3 | 20264621 | 20264622 | 1 | |
| Deletion | Chr12 | 20827807 | 20827829 | 22 | |
| Deletion | Chr7 | 27438459 | 27438498 | 39 | LOC_Os07g45970 |
| Deletion | Chr6 | 28910288 | 28910291 | 3 | LOC_Os06g47770 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22007577 | 22007577 | 1 | |
| Insertion | Chr6 | 9031431 | 9031431 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 12958001 | 13388599 | 2 |
| Inversion | Chr9 | 13234119 | 13307586 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 6153632 | Chr2 | 34427745 | |
| Translocation | Chr11 | 11009197 | Chr10 | 2078898 | LOC_Os10g04400 |
