Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1355-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1355-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 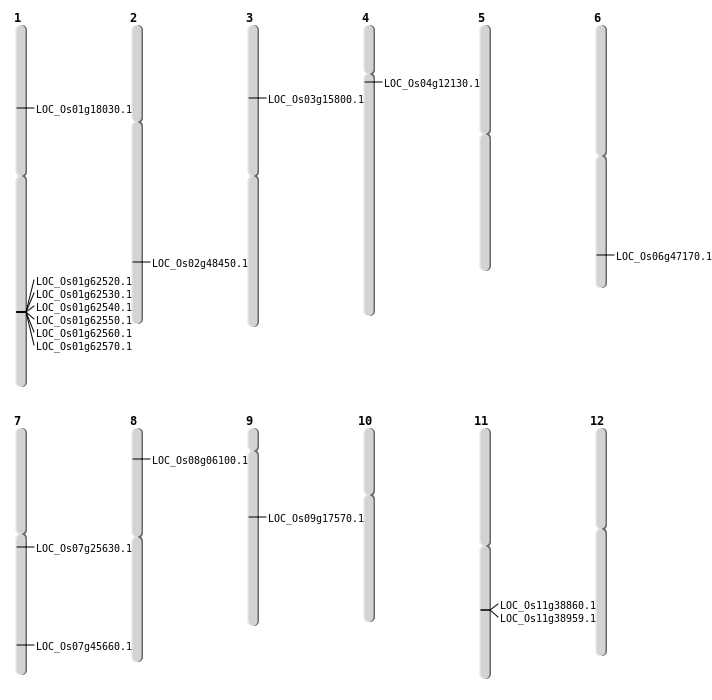
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14091827 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 27168430 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 330104 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1275326 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 17713395 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 23163939 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr11 | 23163943 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr11 | 23219995 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 8106970 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11662984 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 29669388 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g48450 |
| SBS | Chr2 | 29669389 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 31289274 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 34352986 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 7742486 | T-G | HET | INTRON | |
| SBS | Chr4 | 14317018 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 16271157 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 21034841 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 22480475 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 7221917 | A-C | HET | INTRON | |
| SBS | Chr5 | 20092537 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 20287235 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 22345924 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 28359977 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 4248808 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 8136782 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 8136783 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11081380 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 11427970 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 12672722 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17652576 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 28604747 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr6 | 29424944 | G-A | HET | INTRON | |
| SBS | Chr6 | 9566764 | A-C | HET | INTRON | |
| SBS | Chr6 | 9595113 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11774289 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14182528 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 14694096 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25630 |
| SBS | Chr7 | 3265457 | T-A | HOMO | INTRON | |
| SBS | Chr7 | 8437178 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 18837954 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 10746726 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g17570 |
| SBS | Chr9 | 10746727 | C-T | HET | STOP_GAINED | LOC_Os09g17570 |
| SBS | Chr9 | 13805119 | C-G | HOMO | INTRON | |
| SBS | Chr9 | 407376 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7847293 | C-T | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1221493 | 1221505 | 12 | |
| Deletion | Chr8 | 3337774 | 3337785 | 11 | LOC_Os08g06100 |
| Deletion | Chr8 | 4111094 | 4111101 | 7 | |
| Deletion | Chr1 | 4193843 | 4193844 | 1 | |
| Deletion | Chr4 | 6681662 | 6681668 | 6 | LOC_Os04g12130 |
| Deletion | Chr10 | 7788104 | 7788111 | 7 | |
| Deletion | Chr3 | 8719689 | 8719701 | 12 | LOC_Os03g15800 |
| Deletion | Chr1 | 10089136 | 10089141 | 5 | LOC_Os01g18030 |
| Deletion | Chr7 | 10940748 | 10940754 | 6 | |
| Deletion | Chr3 | 11680789 | 11680790 | 1 | |
| Deletion | Chr10 | 15738749 | 15738758 | 9 | |
| Deletion | Chr6 | 15968119 | 15968129 | 10 | |
| Deletion | Chr2 | 18958688 | 18958691 | 3 | |
| Deletion | Chr8 | 19537991 | 19538030 | 39 | |
| Deletion | Chr7 | 21859417 | 21859430 | 13 | |
| Deletion | Chr1 | 23395549 | 23395553 | 4 | |
| Deletion | Chr2 | 23497814 | 23497821 | 7 | |
| Deletion | Chr3 | 25491824 | 25491833 | 9 | |
| Deletion | Chr11 | 25607159 | 25607166 | 7 | |
| Deletion | Chr1 | 26255725 | 26255727 | 2 | |
| Deletion | Chr1 | 26635361 | 26635377 | 16 | |
| Deletion | Chr7 | 27264025 | 27264032 | 7 | LOC_Os07g45660 |
| Deletion | Chr5 | 28859504 | 28859524 | 20 | |
| Deletion | Chr3 | 31092324 | 31092325 | 1 | |
| Deletion | Chr2 | 35138460 | 35138472 | 12 | |
| Deletion | Chr1 | 36203292 | 36224913 | 21621 | 6 |
| Deletion | Chr1 | 42405620 | 42405624 | 4 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 19219927 | 19219930 | 4 | |
| Insertion | Chr4 | 21255083 | 21255083 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 23133377 | 23187455 | 2 |
| Inversion | Chr11 | 23133391 | 23187460 | 2 |
No Translocation
