Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1356-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1356-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 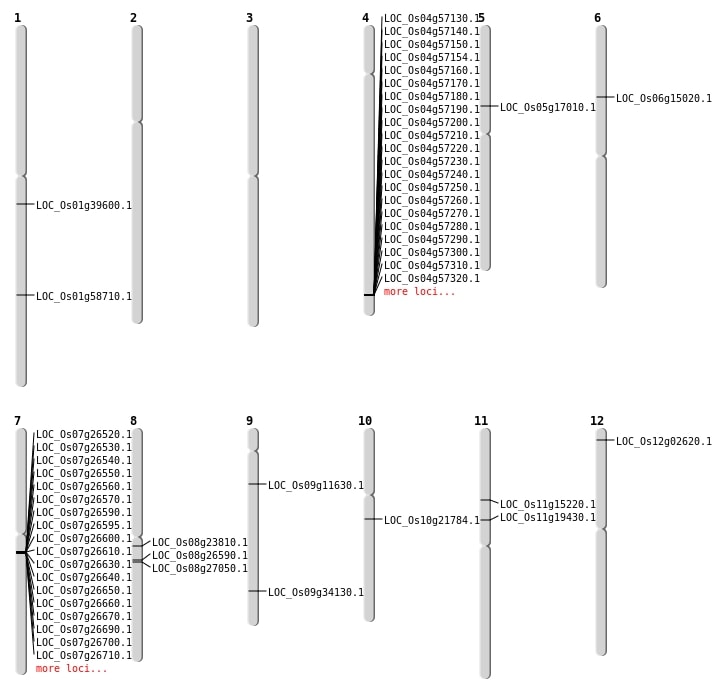
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 33942755 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58710 |
| SBS | Chr1 | 6083720 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12213495 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 3552546 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 936539 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 11177215 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g19430 |
| SBS | Chr11 | 1467932 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16450642 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8940005 | G-C | HET | INTRON | |
| SBS | Chr11 | 9489738 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 1357263 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16523856 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 25907397 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr12 | 908036 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g02620 |
| SBS | Chr12 | 9957165 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 14047343 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 17447746 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20905063 | A-T | HET | INTRON | |
| SBS | Chr2 | 34566539 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 9343768 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 23234876 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 6487413 | G-A | HET | INTRON | |
| SBS | Chr4 | 21432993 | T-A | HET | INTRON | |
| SBS | Chr4 | 35413579 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g59540 |
| SBS | Chr5 | 16266780 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 25818940 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 4198936 | A-T | HET | INTRON | |
| SBS | Chr5 | 8010345 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 25502553 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8511332 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15020 |
| SBS | Chr7 | 17505771 | T-C | HOMO | INTRON | |
| SBS | Chr8 | 12846378 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 21353329 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr9 | 6495531 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11630 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 2791609 | 2791617 | 8 | |
| Deletion | Chr12 | 4813514 | 4813520 | 6 | |
| Deletion | Chr11 | 8590837 | 8590838 | 1 | LOC_Os11g15220 |
| Deletion | Chr7 | 9425931 | 9425933 | 2 | |
| Deletion | Chr5 | 9708262 | 9708263 | 1 | LOC_Os05g17010 |
| Deletion | Chr10 | 11176687 | 11176713 | 26 | LOC_Os10g21784 |
| Deletion | Chr5 | 11682824 | 11682827 | 3 | |
| Deletion | Chr6 | 11722094 | 11722096 | 2 | |
| Deletion | Chr2 | 14745077 | 14745097 | 20 | |
| Deletion | Chr7 | 15280001 | 15468000 | 188000 | 25 |
| Deletion | Chr9 | 17389897 | 17389903 | 6 | |
| Deletion | Chr2 | 17444360 | 17444362 | 2 | |
| Deletion | Chr10 | 17833880 | 17833881 | 1 | |
| Deletion | Chr2 | 20810914 | 20810915 | 1 | |
| Deletion | Chr1 | 22338833 | 22338840 | 7 | LOC_Os01g39600 |
| Deletion | Chr9 | 22440522 | 22440528 | 6 | |
| Deletion | Chr4 | 24064704 | 24064711 | 7 | |
| Deletion | Chr6 | 26230836 | 26230837 | 1 | |
| Deletion | Chr1 | 27436170 | 27436171 | 1 | |
| Deletion | Chr4 | 28905878 | 28905954 | 76 | |
| Deletion | Chr1 | 29017075 | 29017077 | 2 | |
| Deletion | Chr5 | 29714518 | 29714519 | 1 | |
| Deletion | Chr4 | 34035001 | 34449000 | 414000 | 75 |
Insertions: 6
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 16181067 | 16529392 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 14418919 | Chr5 | 7756314 | LOC_Os08g23810 |
| Translocation | Chr9 | 20149350 | Chr8 | 14418941 | 2 |
| Translocation | Chr9 | 20150467 | Chr5 | 7787136 | LOC_Os09g34130 |
