Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1358-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1358-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 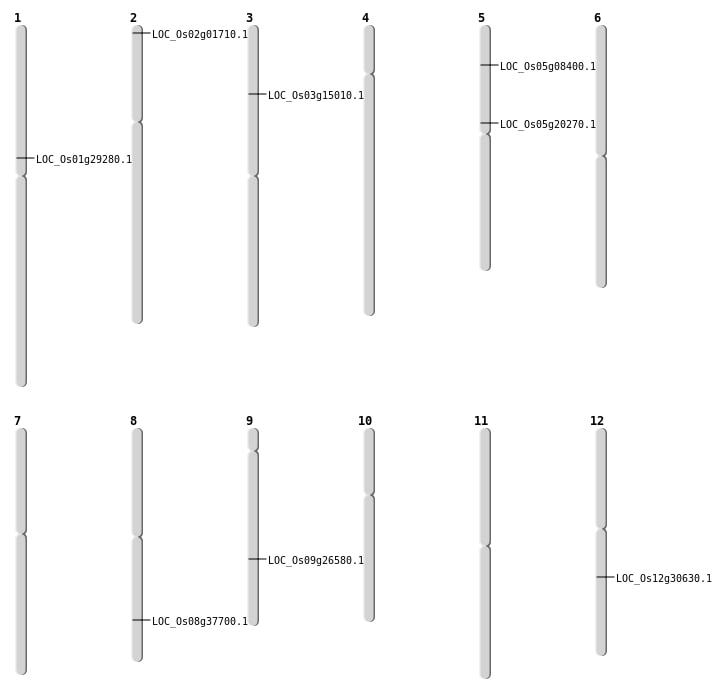
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16128228 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 16415826 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g29280 |
| SBS | Chr1 | 27832972 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 35133823 | G-A | HET | INTRON | |
| SBS | Chr1 | 35934935 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 17162502 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 22328515 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 22426672 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 18395910 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g30630 |
| SBS | Chr12 | 18395912 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 22471676 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 15794830 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 23567489 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 26164165 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr2 | 30059508 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 393225 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g01710 |
| SBS | Chr3 | 14498593 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 23552634 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 13627407 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20519672 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 24724013 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 4582177 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g08400 |
| SBS | Chr6 | 1058053 | C-A | HOMO | INTRON | |
| SBS | Chr6 | 14513067 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 15793288 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 28399553 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 5702720 | T-C | HET | INTRON | |
| SBS | Chr6 | 5716152 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 13931308 | T-C | HOMO | INTRON | |
| SBS | Chr9 | 20008035 | C-A | HOMO | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 263371 | 263373 | 2 | |
| Deletion | Chr7 | 757519 | 757520 | 1 | |
| Deletion | Chr6 | 1022970 | 1023001 | 31 | |
| Deletion | Chr11 | 2099177 | 2099183 | 6 | |
| Deletion | Chr6 | 2344114 | 2344115 | 1 | |
| Deletion | Chr6 | 3030166 | 3030173 | 7 | |
| Deletion | Chr1 | 5852841 | 5852842 | 1 | |
| Deletion | Chr3 | 8193573 | 8193580 | 7 | LOC_Os03g15010 |
| Deletion | Chr12 | 9069876 | 9069890 | 14 | |
| Deletion | Chr10 | 9713150 | 9713153 | 3 | |
| Deletion | Chr2 | 10197302 | 10197307 | 5 | |
| Deletion | Chr10 | 10599120 | 10599125 | 5 | |
| Deletion | Chr7 | 10972017 | 10972019 | 2 | |
| Deletion | Chr5 | 13961581 | 13961587 | 6 | |
| Deletion | Chr3 | 18808367 | 18808377 | 10 | |
| Deletion | Chr6 | 21401799 | 21401819 | 20 | |
| Deletion | Chr2 | 23557841 | 23557866 | 25 | |
| Deletion | Chr8 | 23872596 | 23872601 | 5 | LOC_Os08g37700 |
| Deletion | Chr8 | 24054639 | 24054650 | 11 | |
| Deletion | Chr8 | 26023977 | 26024001 | 24 | |
| Deletion | Chr11 | 26302901 | 26302902 | 1 | |
| Deletion | Chr8 | 28127507 | 28127520 | 13 | |
| Deletion | Chr3 | 29384743 | 29384745 | 2 | |
| Deletion | Chr3 | 29620940 | 29620941 | 1 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 16101764 | 16107552 | LOC_Os09g26580 |
| Inversion | Chr9 | 16101779 | 16107548 | LOC_Os09g26580 |
| Inversion | Chr8 | 22708344 | 22781481 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 11877870 | Chr1 | 36025161 | LOC_Os05g20270 |
| Translocation | Chr5 | 11879119 | Chr1 | 36025157 |
