Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1363-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1363-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 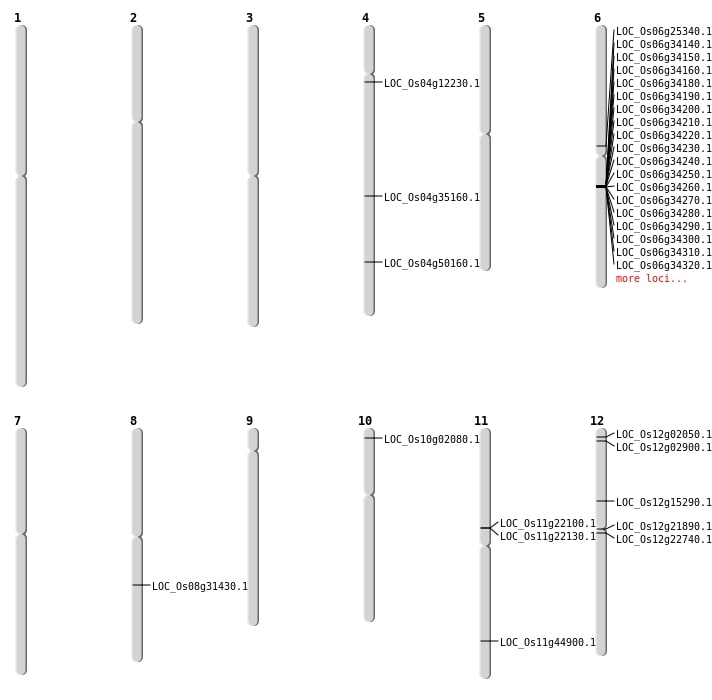
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18807870 | G-A | HET | INTRON | |
| SBS | Chr1 | 26418162 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 4297411 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 680632 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g02080 |
| SBS | Chr11 | 10416271 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 12689574 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22100 |
| SBS | Chr11 | 16973509 | G-A | HET | INTRON | |
| SBS | Chr11 | 8522325 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 10273687 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 10273688 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 1063972 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g02900 |
| SBS | Chr12 | 12328796 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g21890 |
| SBS | Chr12 | 8721860 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15290 |
| SBS | Chr2 | 18699763 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 21777484 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 25380231 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 27570091 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 16622103 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 35302567 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 6746436 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12230 |
| SBS | Chr5 | 21584408 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 3736958 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 14892632 | C-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 406659 | G-A | HET | INTRON | |
| SBS | Chr6 | 4202179 | A-G | HET | INTRON | |
| SBS | Chr7 | 2650189 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 28586049 | T-G | HET | INTRON | |
| SBS | Chr8 | 11393953 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 12618391 | G-A | HET | INTRON | |
| SBS | Chr8 | 2398916 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 27746761 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 14945715 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 16167597 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 6901690 | A-T | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 44819 | 44820 | 1 | |
| Deletion | Chr3 | 326087 | 326098 | 11 | |
| Deletion | Chr1 | 1450632 | 1450637 | 5 | |
| Deletion | Chr12 | 4060413 | 4060414 | 1 | |
| Deletion | Chr7 | 4713679 | 4713685 | 6 | |
| Deletion | Chr8 | 5680495 | 5680498 | 3 | |
| Deletion | Chr7 | 6188648 | 6188649 | 1 | |
| Deletion | Chr5 | 10642730 | 10642738 | 8 | |
| Deletion | Chr12 | 12829128 | 12829153 | 25 | LOC_Os12g22740 |
| Deletion | Chr6 | 14841969 | 14841970 | 1 | LOC_Os06g25340 |
| Deletion | Chr1 | 14902200 | 14902201 | 1 | |
| Deletion | Chr11 | 16981591 | 16981620 | 29 | |
| Deletion | Chr8 | 19441395 | 19441396 | 1 | LOC_Os08g31430 |
| Deletion | Chr6 | 19875542 | 19977309 | 101767 | 19 |
| Deletion | Chr10 | 20191874 | 20191875 | 1 | |
| Deletion | Chr4 | 21370542 | 21370549 | 7 | LOC_Os04g35160 |
| Deletion | Chr11 | 21605899 | 21605911 | 12 | |
| Deletion | Chr1 | 23934596 | 23934606 | 10 | |
| Deletion | Chr5 | 24426044 | 24426061 | 17 | |
| Deletion | Chr6 | 28273019 | 28273027 | 8 | |
| Deletion | Chr4 | 29916843 | 29916862 | 19 | LOC_Os04g50160 |
| Deletion | Chr1 | 37434690 | 37434696 | 6 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 27021465 | 27176319 | LOC_Os11g44900 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 610774 | Chr4 | 4507458 | LOC_Os12g02050 |
| Translocation | Chr11 | 12699509 | Chr7 | 14859316 | LOC_Os11g22130 |
