Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1366-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1366-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 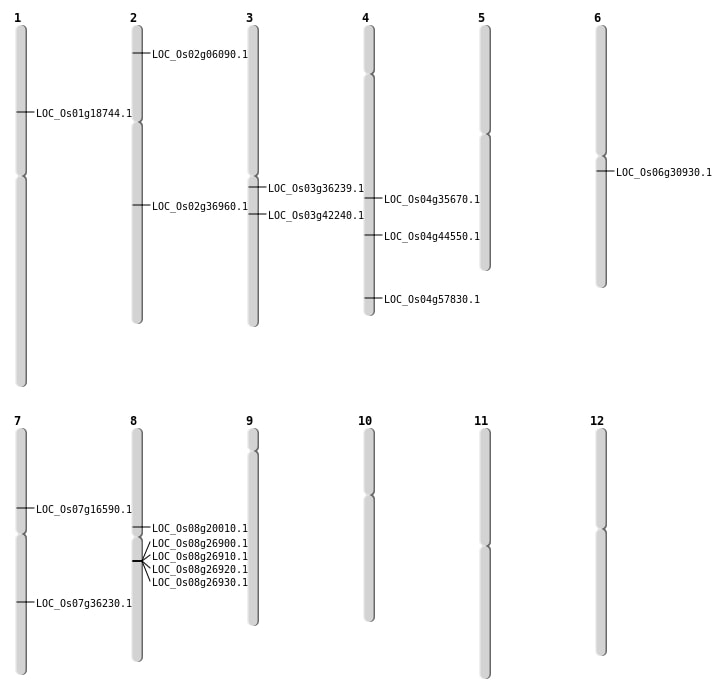
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10565053 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g18744 |
| SBS | Chr1 | 12513187 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 1255994 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 1849064 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 21428496 | C-G | HOMO | INTRON | |
| SBS | Chr1 | 35555869 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 40765469 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 9742406 | T-A | HOMO | INTRON | |
| SBS | Chr10 | 14324351 | T-A | HET | INTRON | |
| SBS | Chr10 | 17017664 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 20155646 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 10943526 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 13639822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 14009492 | G-A | HET | INTRON | |
| SBS | Chr2 | 22327773 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g36960 |
| SBS | Chr2 | 30821913 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 5283200 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 6826952 | A-G | HET | INTRON | |
| SBS | Chr2 | 6830149 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 8296498 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 20845672 | C-T | HET | INTRON | |
| SBS | Chr3 | 23500503 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g42240 |
| SBS | Chr3 | 3623580 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 26849222 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 3077745 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 3574889 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 8184388 | G-A | HET | INTRON | |
| SBS | Chr5 | 15305393 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 19784657 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr5 | 20656742 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 21273092 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 3270344 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 3271068 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 5435539 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 16359953 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 16822381 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 17039820 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 17039821 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 7797539 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 20767926 | G-A | HET | INTRON | |
| SBS | Chr7 | 23815365 | T-G | HET | INTRON | |
| SBS | Chr7 | 8867456 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 9730111 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16590 |
| SBS | Chr8 | 20521428 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 21517927 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 3316852 | C-A | HOMO | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 974023 | 974024 | 1 | |
| Deletion | Chr2 | 3038135 | 3038144 | 9 | LOC_Os02g06090 |
| Deletion | Chr5 | 3101363 | 3101369 | 6 | |
| Deletion | Chr3 | 3430196 | 3430200 | 4 | |
| Deletion | Chr5 | 5822986 | 5822999 | 13 | |
| Deletion | Chr5 | 7336336 | 7336337 | 1 | |
| Deletion | Chr12 | 8125517 | 8125532 | 15 | |
| Deletion | Chr8 | 11441025 | 11441026 | 1 | |
| Deletion | Chr8 | 11983579 | 11983597 | 18 | LOC_Os08g20010 |
| Deletion | Chr10 | 12957131 | 12957132 | 1 | |
| Deletion | Chr6 | 14242065 | 14242070 | 5 | |
| Deletion | Chr8 | 16403001 | 16434000 | 31000 | 4 |
| Deletion | Chr12 | 16898852 | 16898864 | 12 | |
| Deletion | Chr6 | 17995029 | 17995041 | 12 | LOC_Os06g30930 |
| Deletion | Chr8 | 19248908 | 19248909 | 1 | |
| Deletion | Chr3 | 20106412 | 20106413 | 1 | LOC_Os03g36239 |
| Deletion | Chr4 | 21739134 | 21739135 | 1 | LOC_Os04g35670 |
| Deletion | Chr7 | 21848472 | 21848496 | 24 | |
| Deletion | Chr4 | 22002222 | 22002223 | 1 | |
| Deletion | Chr7 | 22310134 | 22310175 | 41 | |
| Deletion | Chr8 | 22899437 | 22899438 | 1 | |
| Deletion | Chr7 | 23673891 | 23673928 | 37 | |
| Deletion | Chr8 | 23883710 | 23883712 | 2 | |
| Deletion | Chr8 | 23954295 | 23954304 | 9 | |
| Deletion | Chr4 | 26374767 | 26374770 | 3 | LOC_Os04g44550 |
| Deletion | Chr2 | 27196439 | 27196453 | 14 | |
| Deletion | Chr2 | 32627697 | 32627699 | 2 | |
| Deletion | Chr4 | 34437373 | 34437384 | 11 | LOC_Os04g57830 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 28901005 | 28901005 | 1 | |
| Insertion | Chr6 | 7530175 | 7530175 | 1 | |
| Insertion | Chr9 | 5020072 | 5020073 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 16993323 | 17553464 | |
| Inversion | Chr7 | 20693966 | 21650853 | |
| Inversion | Chr7 | 20693981 | 21650852 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 11009299 | Chr10 | 2076721 | |
| Translocation | Chr7 | 21657366 | Chr4 | 31867777 | LOC_Os07g36230 |
