Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1369-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1369-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 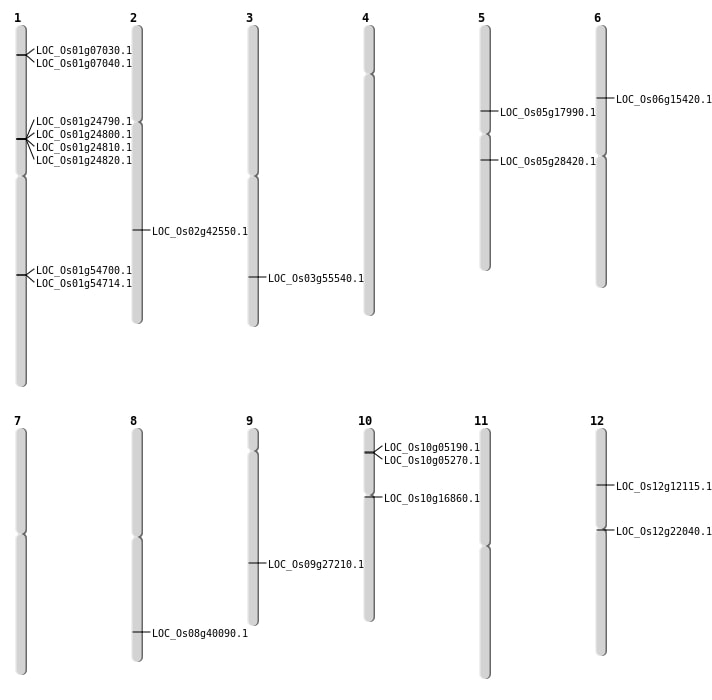
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18796985 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 24031522 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 42289718 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 1071114 | A-T | Homo | INTRON | |
| SBS | Chr10 | 8401433 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g16860 |
| SBS | Chr2 | 15097934 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 26429431 | A-G | HET | INTRON | |
| SBS | Chr2 | 28197941 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 30670263 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 3345412 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 4264734 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 7257229 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 8751426 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 13241215 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1605980 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 19045630 | C-T | HET | INTRON | |
| SBS | Chr5 | 10340717 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17990 |
| SBS | Chr5 | 16640887 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g28420 |
| SBS | Chr5 | 20369899 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 5740579 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8503965 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 22414984 | T-A | Homo | UTR_3_PRIME | |
| SBS | Chr8 | 23245283 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 3558247 | A-G | Homo | INTERGENIC | |
| SBS | Chr9 | 10489313 | T-G | Homo | INTRON | |
| SBS | Chr9 | 924126 | A-G | Homo | INTRON |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1150238 | 1150239 | 1 | |
| Deletion | Chr9 | 3039269 | 3039271 | 2 | |
| Deletion | Chr1 | 3312974 | 3315306 | 2332 | 2 |
| Deletion | Chr2 | 3710844 | 3710845 | 1 | |
| Deletion | Chr3 | 5203978 | 5203983 | 5 | |
| Deletion | Chr6 | 6326105 | 6326127 | 22 | |
| Deletion | Chr6 | 6338076 | 6338077 | 1 | |
| Deletion | Chr12 | 6643334 | 6643547 | 213 | LOC_Os12g12115 |
| Deletion | Chr12 | 8380055 | 8380059 | 4 | |
| Deletion | Chr9 | 9199067 | 9199068 | 1 | |
| Deletion | Chr2 | 9909052 | 9909053 | 1 | |
| Deletion | Chr6 | 12203800 | 12203801 | 1 | |
| Deletion | Chr6 | 12361837 | 12361838 | 1 | |
| Deletion | Chr7 | 12413923 | 12413928 | 5 | |
| Deletion | Chr12 | 12694284 | 12694285 | 1 | |
| Deletion | Chr1 | 13954001 | 13980000 | 26000 | 4 |
| Deletion | Chr4 | 14803105 | 14803106 | 1 | |
| Deletion | Chr7 | 14854529 | 14854532 | 3 | |
| Deletion | Chr8 | 17449121 | 17449122 | 1 | |
| Deletion | Chr6 | 18087954 | 18087955 | 1 | |
| Deletion | Chr6 | 19510936 | 19510942 | 6 | |
| Deletion | Chr5 | 22222089 | 22222090 | 1 | |
| Deletion | Chr2 | 22412606 | 22412610 | 4 | |
| Deletion | Chr1 | 22533612 | 22533616 | 4 | |
| Deletion | Chr3 | 23096682 | 23096700 | 18 | |
| Deletion | Chr8 | 23328525 | 23328526 | 1 | |
| Deletion | Chr2 | 23738233 | 23738234 | 1 | |
| Deletion | Chr12 | 24181333 | 24181648 | 315 | |
| Deletion | Chr5 | 25413702 | 25413712 | 10 | |
| Deletion | Chr5 | 27180711 | 27180712 | 1 | |
| Deletion | Chr1 | 31076448 | 31076451 | 3 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 22375831 | 22375831 | 1 | |
| Insertion | Chr10 | 2543900 | 2543932 | 33 | LOC_Os10g05190 |
| Insertion | Chr10 | 2607012 | 2607074 | 63 | LOC_Os10g05270 |
| Insertion | Chr12 | 25845451 | 25845464 | 14 | |
| Insertion | Chr12 | 3661338 | 3661339 | 2 | |
| Insertion | Chr2 | 34436552 | 34436552 | 1 | |
| Insertion | Chr2 | 7749768 | 7749769 | 2 | |
| Insertion | Chr3 | 31605866 | 31605867 | 2 | LOC_Os03g55540 |
| Insertion | Chr4 | 20356376 | 20356377 | 2 | |
| Insertion | Chr5 | 24126885 | 24126885 | 1 | |
| Insertion | Chr5 | 733095 | 733096 | 2 | |
| Insertion | Chr6 | 18308189 | 18308189 | 1 | |
| Insertion | Chr6 | 20840493 | 20840493 | 1 | |
| Insertion | Chr6 | 4833590 | 4833593 | 4 | |
| Insertion | Chr8 | 20286337 | 20286340 | 4 | |
| Insertion | Chr8 | 25381036 | 25381041 | 6 | LOC_Os08g40090 |
| Insertion | Chr9 | 4642134 | 4642134 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 6643338 | 20105659 | |
| Inversion | Chr12 | 6643528 | 21555787 | LOC_Os12g12115 |
| Inversion | Chr6 | 7687437 | 8759926 | LOC_Os06g15420 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 934980 | Chr6 | 7687436 | |
| Translocation | Chr9 | 956564 | Chr6 | 8759966 | LOC_Os06g15420 |
| Translocation | Chr12 | 12408507 | Chr9 | 16546853 | 2 |
| Translocation | Chr2 | 25580913 | Chr1 | 31477562 | 2 |
| Translocation | Chr2 | 25580924 | Chr1 | 31482662 | 2 |
