Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1371-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1371-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 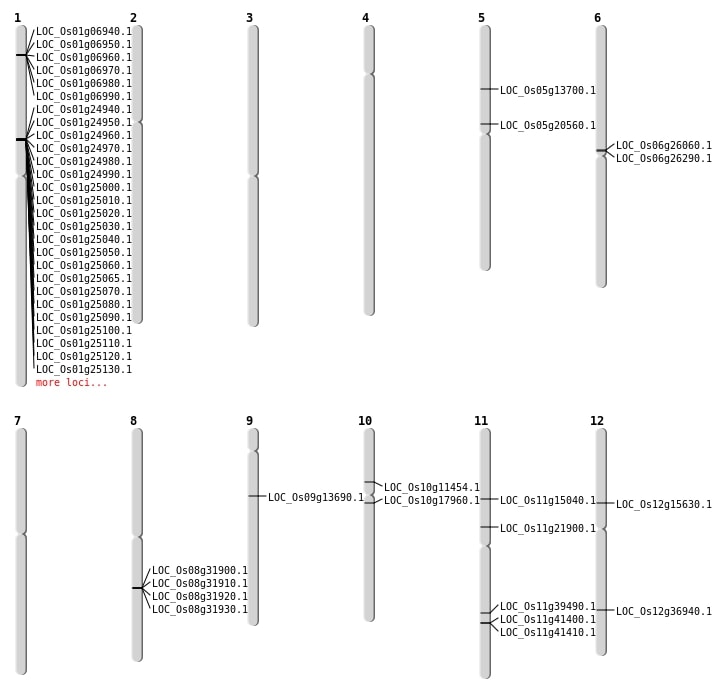
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15527524 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g27800 |
| SBS | Chr1 | 19503162 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 25385238 | T-A | Homo | INTERGENIC | |
| SBS | Chr1 | 26418162 | G-T | Homo | INTERGENIC | |
| SBS | Chr1 | 37102784 | G-A | Homo | INTERGENIC | |
| SBS | Chr1 | 38970513 | A-G | Homo | INTERGENIC | |
| SBS | Chr1 | 5011172 | C-T | HET | INTRON | |
| SBS | Chr10 | 12494393 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 6340150 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g11454 |
| SBS | Chr11 | 18023555 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 2349392 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 28863297 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 7538600 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 16259786 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 7204675 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8925070 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15630 |
| SBS | Chr2 | 10112219 | C-A | Homo | INTRON | |
| SBS | Chr2 | 13300538 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10188230 | A-G | HET | INTRON | |
| SBS | Chr3 | 20236885 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 15463632 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 30973737 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 4967795 | G-A | Homo | INTRON | |
| SBS | Chr4 | 9870856 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 18516988 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 26551368 | C-A | HET | INTRON | |
| SBS | Chr5 | 7592009 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g13700 |
| SBS | Chr5 | 7863982 | A-T | Homo | INTERGENIC | |
| SBS | Chr6 | 12773057 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 12773058 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 22461145 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 9240332 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 10503253 | A-G | Homo | INTERGENIC | |
| SBS | Chr8 | 27746761 | C-T | Homo | INTERGENIC | |
| SBS | Chr9 | 12249261 | A-G | Homo | INTERGENIC | |
| SBS | Chr9 | 8008084 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g13690 |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2278684 | 2278698 | 14 | |
| Deletion | Chr10 | 3170499 | 3170501 | 2 | |
| Deletion | Chr1 | 3288486 | 3298013 | 9527 | 6 |
| Deletion | Chr2 | 3332805 | 3332806 | 1 | |
| Deletion | Chr12 | 3424152 | 3424159 | 7 | |
| Deletion | Chr4 | 4964219 | 4964224 | 5 | |
| Deletion | Chr11 | 8467816 | 8468144 | 328 | LOC_Os11g15040 |
| Deletion | Chr10 | 9104745 | 9104746 | 1 | LOC_Os10g17960 |
| Deletion | Chr6 | 10108926 | 10108932 | 6 | |
| Deletion | Chr12 | 11064522 | 11064525 | 3 | |
| Deletion | Chr11 | 11107384 | 11107385 | 1 | |
| Deletion | Chr3 | 11386449 | 11386450 | 1 | |
| Deletion | Chr10 | 11557482 | 11557484 | 2 | |
| Deletion | Chr1 | 12224509 | 12224511 | 2 | |
| Deletion | Chr11 | 12573407 | 12573422 | 15 | LOC_Os11g21900 |
| Deletion | Chr1 | 14054001 | 14214000 | 160000 | 24 |
| Deletion | Chr11 | 14518650 | 14518652 | 2 | |
| Deletion | Chr10 | 17353426 | 17353475 | 49 | |
| Deletion | Chr4 | 18751574 | 18751583 | 9 | |
| Deletion | Chr11 | 18830529 | 18830536 | 7 | |
| Deletion | Chr4 | 19439565 | 19439567 | 2 | |
| Deletion | Chr2 | 19491370 | 19491372 | 2 | |
| Deletion | Chr5 | 19603037 | 19603052 | 15 | |
| Deletion | Chr8 | 19768001 | 19798000 | 30000 | 4 |
| Deletion | Chr8 | 21660278 | 21660283 | 5 | |
| Deletion | Chr7 | 22199943 | 22199955 | 12 | |
| Deletion | Chr12 | 22653567 | 22653568 | 1 | LOC_Os12g36940 |
| Deletion | Chr11 | 23512377 | 23512392 | 15 | LOC_Os11g39490 |
| Deletion | Chr4 | 23587595 | 23587599 | 4 | |
| Deletion | Chr8 | 23844924 | 23844950 | 26 | |
| Deletion | Chr11 | 24843990 | 24852538 | 8548 | 2 |
| Deletion | Chr11 | 25011216 | 25011224 | 8 | |
| Deletion | Chr1 | 25354687 | 25354702 | 15 | |
| Deletion | Chr4 | 28516837 | 28516847 | 10 | |
| Deletion | Chr2 | 30844068 | 30844081 | 13 | |
| Deletion | Chr2 | 31062462 | 31062464 | 2 | |
| Deletion | Chr4 | 32334474 | 32334771 | 297 | |
| Deletion | Chr2 | 35246512 | 35246518 | 6 |
Insertions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 13262373 | 13262416 | 44 | |
| Insertion | Chr11 | 15729242 | 15729294 | 53 | |
| Insertion | Chr11 | 22670209 | 22670284 | 76 | |
| Insertion | Chr11 | 23031411 | 23031413 | 3 | |
| Insertion | Chr11 | 27392656 | 27392666 | 11 | |
| Insertion | Chr11 | 28218065 | 28218065 | 1 | |
| Insertion | Chr12 | 10496638 | 10496639 | 2 | |
| Insertion | Chr12 | 13016242 | 13016242 | 1 | |
| Insertion | Chr2 | 9264875 | 9264875 | 1 | |
| Insertion | Chr4 | 23681155 | 23681155 | 1 | |
| Insertion | Chr5 | 12064532 | 12064532 | 1 | LOC_Os05g20560 |
| Insertion | Chr7 | 25028812 | 25028812 | 1 | |
| Insertion | Chr8 | 14844730 | 14844731 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 15248943 | 15392062 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 8009749 | Chr2 | 10396049 |
