Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1373-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1373-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 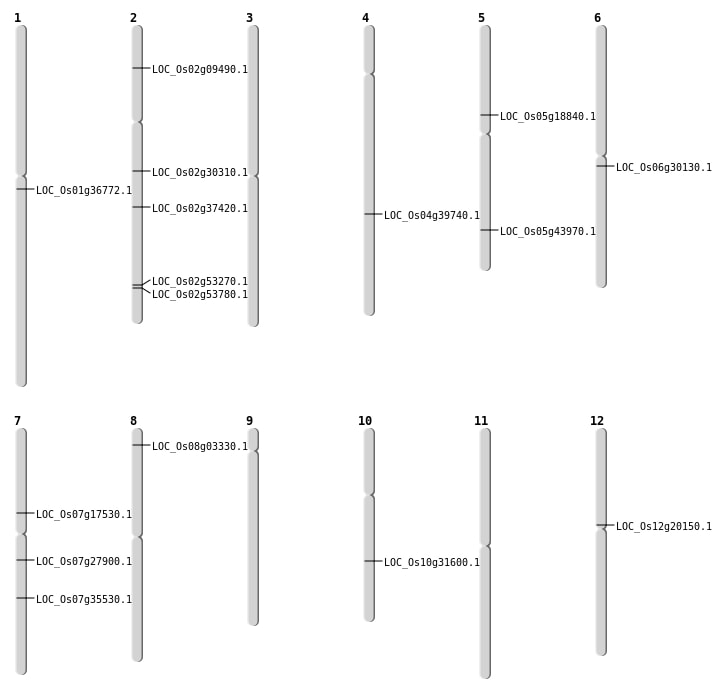
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 16560729 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g31600 |
| SBS | Chr10 | 4319101 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 10244442 | G-A | Homo | INTRON | |
| SBS | Chr11 | 9543104 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 18046315 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g30310 |
| SBS | Chr2 | 19115990 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 22014209 | C-T | HET | INTRON | |
| SBS | Chr2 | 35716447 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 4870390 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g09490 |
| SBS | Chr3 | 23886664 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 14668467 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21050234 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 10943016 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g18840 |
| SBS | Chr5 | 1965199 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 24448941 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 16027038 | G-A | HET | INTRON | |
| SBS | Chr6 | 21062220 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 21938071 | G-A | HET | INTRON | |
| SBS | Chr6 | 8949199 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17696803 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 21263949 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g35530 |
| SBS | Chr7 | 9162061 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 11115005 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 1544448 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g03330 |
| SBS | Chr8 | 17866939 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 17866940 | T-A | Homo | INTERGENIC | |
| SBS | Chr8 | 27624035 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 4727519 | C-G | Homo | INTERGENIC | |
| SBS | Chr8 | 8449517 | C-T | Homo | INTRON | |
| SBS | Chr9 | 1621678 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 61921 | G-A | Homo | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 3884069 | 3884072 | 3 | |
| Deletion | Chr1 | 5687246 | 5687255 | 9 | |
| Deletion | Chr7 | 7071265 | 7071269 | 4 | |
| Deletion | Chr6 | 8096042 | 8096043 | 1 | |
| Deletion | Chr12 | 9971137 | 9971153 | 16 | |
| Deletion | Chr10 | 10314976 | 10314979 | 3 | |
| Deletion | Chr7 | 10360149 | 10360163 | 14 | |
| Deletion | Chr7 | 10361929 | 10361930 | 1 | LOC_Os07g17530 |
| Deletion | Chr3 | 10951888 | 10951893 | 5 | |
| Deletion | Chr8 | 11658342 | 11658343 | 1 | |
| Deletion | Chr12 | 11727859 | 11727867 | 8 | LOC_Os12g20150 |
| Deletion | Chr10 | 13603270 | 13603271 | 1 | |
| Deletion | Chr9 | 13724000 | 13724004 | 4 | |
| Deletion | Chr7 | 16278764 | 16278765 | 1 | LOC_Os07g27900 |
| Deletion | Chr6 | 17379356 | 17379357 | 1 | LOC_Os06g30130 |
| Deletion | Chr11 | 17976010 | 17976014 | 4 | |
| Deletion | Chr7 | 18709609 | 18709620 | 11 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr1 | 20429513 | 20429531 | 18 | LOC_Os01g36772 |
| Deletion | Chr6 | 21062243 | 21062244 | 1 | |
| Deletion | Chr3 | 21284141 | 21284149 | 8 | |
| Deletion | Chr10 | 21963548 | 21963549 | 1 | |
| Deletion | Chr4 | 23683489 | 23683493 | 4 | LOC_Os04g39740 |
| Deletion | Chr2 | 29644711 | 29644712 | 1 | |
| Deletion | Chr1 | 31282223 | 31282224 | 1 | |
| Deletion | Chr2 | 32947313 | 32947358 | 45 | LOC_Os02g53780 |
Insertions: 7
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 22600869 | 22685197 | LOC_Os02g37420 |
| Inversion | Chr2 | 32337327 | 32608431 | LOC_Os02g53270 |
| Inversion | Chr2 | 32337340 | 33999464 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 25582593 | Chr2 | 33998471 | LOC_Os05g43970 |
