Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1384-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1384-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 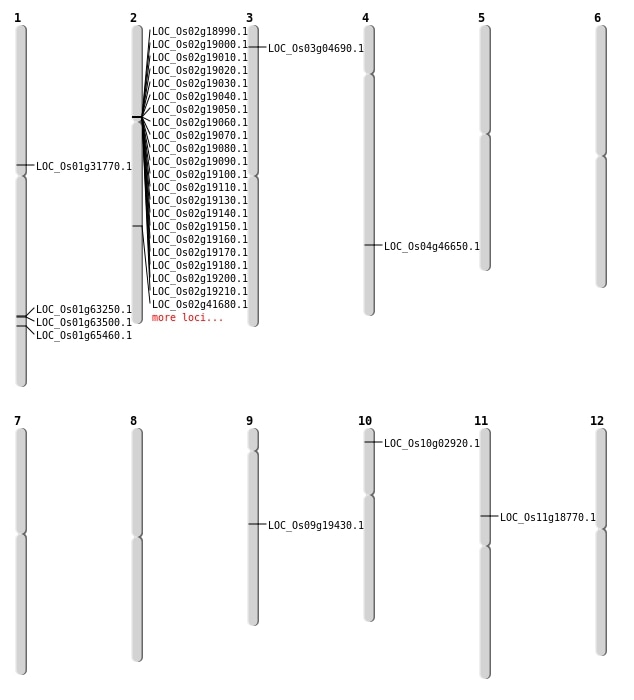
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17399669 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g31770 |
| SBS | Chr1 | 24657517 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 27934232 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 36791686 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 37005819 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 38498349 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40366243 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 7991525 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 1191786 | C-A | HET | START_LOST | LOC_Os10g02920 |
| SBS | Chr10 | 22382694 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11179070 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 27294477 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11578327 | G-A | HET | INTRON | |
| SBS | Chr12 | 2765833 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 710254 | T-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 11172287 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 14880048 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 24514837 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 8715098 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 2163548 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 32490353 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 27656234 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g46650 |
| SBS | Chr5 | 16890867 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 28496345 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 28496348 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 4626375 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 14026251 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 20866383 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 3075204 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 6656326 | T-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 28130065 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 5127276 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 5127277 | C-T | HOMO | INTRON | |
| SBS | Chr9 | 11627114 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19430 |
| SBS | Chr9 | 14584814 | C-T | HET | INTRON | |
| SBS | Chr9 | 15279837 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 17658022 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 9846215 | G-A | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 549866 | 549867 | 1 | |
| Deletion | Chr10 | 1847351 | 1847368 | 17 | |
| Deletion | Chr3 | 2225677 | 2225680 | 3 | LOC_Os03g04690 |
| Deletion | Chr1 | 3407676 | 3407684 | 8 | |
| Deletion | Chr3 | 3412140 | 3412141 | 1 | |
| Deletion | Chr2 | 7736034 | 7736036 | 2 | |
| Deletion | Chr1 | 9072990 | 9072991 | 1 | |
| Deletion | Chr2 | 11095001 | 11204000 | 109000 | 21 |
| Deletion | Chr3 | 12727113 | 12727117 | 4 | |
| Deletion | Chr11 | 13940256 | 13940257 | 1 | |
| Deletion | Chr2 | 14155098 | 14155105 | 7 | |
| Deletion | Chr7 | 14761353 | 14761354 | 1 | |
| Deletion | Chr1 | 15175542 | 15175551 | 9 | |
| Deletion | Chr3 | 15851041 | 15851042 | 1 | |
| Deletion | Chr10 | 16888601 | 16888614 | 13 | |
| Deletion | Chr1 | 17450947 | 17450949 | 2 | |
| Deletion | Chr5 | 22740433 | 22740435 | 2 | |
| Deletion | Chr2 | 22996180 | 22996184 | 4 | |
| Deletion | Chr1 | 24114378 | 24114388 | 10 | |
| Deletion | Chr2 | 25008883 | 25008884 | 1 | LOC_Os02g41680 |
| Deletion | Chr11 | 28926058 | 28926066 | 8 | |
| Deletion | Chr1 | 37999652 | 37999670 | 18 | LOC_Os01g65460 |
Insertions: 7
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 36660649 | 36808776 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 10654925 | Chr2 | 31786583 | 2 |
| Translocation | Chr11 | 10656206 | Chr2 | 31786575 | 2 |
| Translocation | Chr11 | 10656209 | Chr3 | 11091592 | LOC_Os11g18770 |
| Translocation | Chr11 | 10656467 | Chr3 | 11091573 | LOC_Os11g18770 |
