Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1389-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1389-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 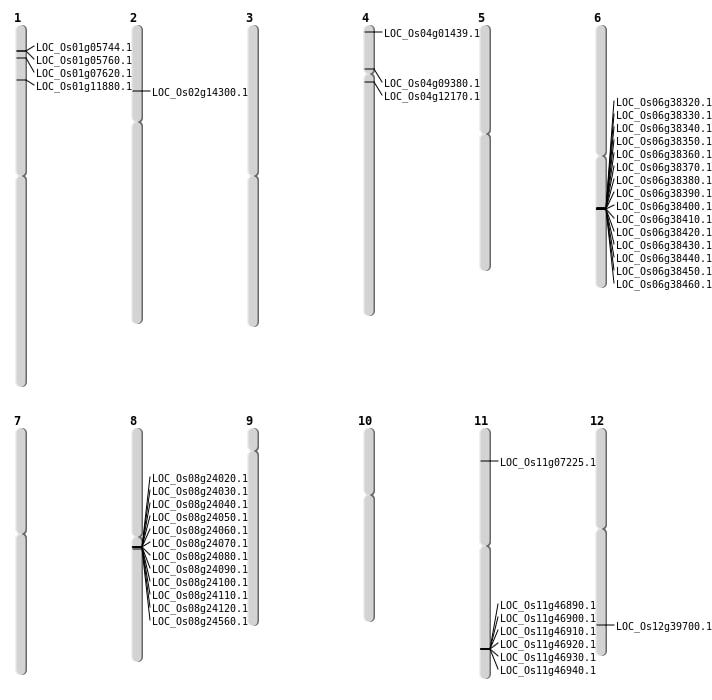
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12651770 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr1 | 32526290 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr1 | 9887308 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 10455174 | C-A | Homo | INTERGENIC | |
| SBS | Chr10 | 14396361 | C-T | HET | INTRON | |
| SBS | Chr10 | 19592260 | C-G | Homo | INTERGENIC | |
| SBS | Chr10 | 20709105 | T-G | Homo | INTERGENIC | |
| SBS | Chr10 | 22005794 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 27765419 | G-A | HET | INTRON | |
| SBS | Chr11 | 3630424 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g07225 |
| SBS | Chr12 | 13210823 | C-T | Homo | INTERGENIC | |
| SBS | Chr12 | 15459930 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 15607494 | T-C | HET | INTRON | |
| SBS | Chr12 | 6893438 | G-T | Homo | INTRON | |
| SBS | Chr2 | 33307238 | A-G | HET | INTRON | |
| SBS | Chr3 | 21991446 | C-G | HET | INTRON | |
| SBS | Chr3 | 25222710 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 35136987 | G-A | Homo | UTR_5_PRIME | |
| SBS | Chr3 | 36031191 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 18124105 | T-G | HET | INTRON | |
| SBS | Chr4 | 34955200 | A-G | Homo | INTERGENIC | |
| SBS | Chr4 | 6706868 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12170 |
| SBS | Chr5 | 12112153 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 14830369 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 15300018 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 13246105 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 14543329 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 16596929 | T-G | Homo | INTRON | |
| SBS | Chr7 | 10458626 | G-T | HET | INTRON | |
| SBS | Chr7 | 14088285 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 14088451 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 17016057 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 2542818 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 721232 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 863364 | T-A | HET | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 175507 | 175509 | 2 | |
| Deletion | Chr4 | 298915 | 309262 | 10347 | LOC_Os04g01439 |
| Deletion | Chr1 | 809421 | 809424 | 3 | |
| Deletion | Chr11 | 2683793 | 2683798 | 5 | |
| Deletion | Chr1 | 2743398 | 2752975 | 9577 | 2 |
| Deletion | Chr1 | 3665198 | 3665202 | 4 | LOC_Os01g07620 |
| Deletion | Chr3 | 4552256 | 4552257 | 1 | |
| Deletion | Chr1 | 6432555 | 6432556 | 1 | LOC_Os01g11880 |
| Deletion | Chr8 | 7266353 | 7266361 | 8 | |
| Deletion | Chr3 | 7439450 | 7439452 | 2 | |
| Deletion | Chr5 | 8235960 | 8235962 | 2 | |
| Deletion | Chr1 | 9797899 | 9797902 | 3 | |
| Deletion | Chr12 | 12051331 | 12051332 | 1 | |
| Deletion | Chr7 | 12427752 | 12427756 | 4 | |
| Deletion | Chr7 | 13504503 | 13504532 | 29 | |
| Deletion | Chr8 | 14524001 | 14569000 | 45000 | 11 |
| Deletion | Chr8 | 16922091 | 16922092 | 1 | |
| Deletion | Chr1 | 17436990 | 17436995 | 5 | |
| Deletion | Chr6 | 18791046 | 18791049 | 3 | |
| Deletion | Chr1 | 20455045 | 20455058 | 13 | |
| Deletion | Chr7 | 21165365 | 21165370 | 5 | |
| Deletion | Chr5 | 22300882 | 22300884 | 2 | |
| Deletion | Chr12 | 22471189 | 22471190 | 1 | |
| Deletion | Chr6 | 22687001 | 22794000 | 107000 | 15 |
| Deletion | Chr6 | 23895267 | 23895268 | 1 | |
| Deletion | Chr12 | 24525460 | 24525462 | 2 | LOC_Os12g39700 |
| Deletion | Chr8 | 26079252 | 26079253 | 1 | |
| Deletion | Chr11 | 26714589 | 26714590 | 1 | |
| Deletion | Chr12 | 27291929 | 27291930 | 1 | |
| Deletion | Chr11 | 28172001 | 28211000 | 39000 | 6 |
| Deletion | Chr3 | 29383315 | 29383419 | 104 | |
| Deletion | Chr1 | 29715567 | 29715568 | 1 | |
| Deletion | Chr6 | 30787733 | 30787757 | 24 | |
| Deletion | Chr2 | 34070556 | 34070557 | 1 | |
| Deletion | Chr1 | 40899863 | 40899867 | 4 | |
| Deletion | Chr1 | 41690782 | 41690792 | 10 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11052183 | 11052184 | 2 | |
| Insertion | Chr1 | 14871061 | 14871065 | 5 | |
| Insertion | Chr1 | 16209500 | 16209500 | 1 | |
| Insertion | Chr1 | 2285352 | 2285352 | 1 | |
| Insertion | Chr10 | 7138490 | 7138490 | 1 | |
| Insertion | Chr11 | 10895601 | 10895601 | 1 | |
| Insertion | Chr11 | 11652580 | 11652582 | 3 | |
| Insertion | Chr11 | 12120993 | 12121045 | 53 | |
| Insertion | Chr2 | 34287786 | 34287789 | 4 | |
| Insertion | Chr3 | 30494350 | 30494350 | 1 | |
| Insertion | Chr4 | 20974079 | 20974086 | 8 | |
| Insertion | Chr5 | 21451997 | 21451998 | 2 | |
| Insertion | Chr5 | 3916730 | 3916730 | 1 | |
| Insertion | Chr5 | 8933206 | 8933206 | 1 | |
| Insertion | Chr6 | 5718480 | 5718481 | 2 | |
| Insertion | Chr7 | 27282369 | 27282370 | 2 | |
| Insertion | Chr8 | 14841825 | 14841825 | 1 | LOC_Os08g24560 |
| Insertion | Chr8 | 28276174 | 28276175 | 2 | |
| Insertion | Chr9 | 5009001 | 5009004 | 4 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6632107 | 6662292 | |
| Inversion | Chr2 | 7838671 | 7906221 | |
| Inversion | Chr2 | 7853090 | 7906222 | LOC_Os02g14300 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 17250533 | Chr4 | 5000289 | LOC_Os04g09380 |
| Translocation | Chr11 | 17492770 | Chr3 | 25243467 |
