Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1392-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1392-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 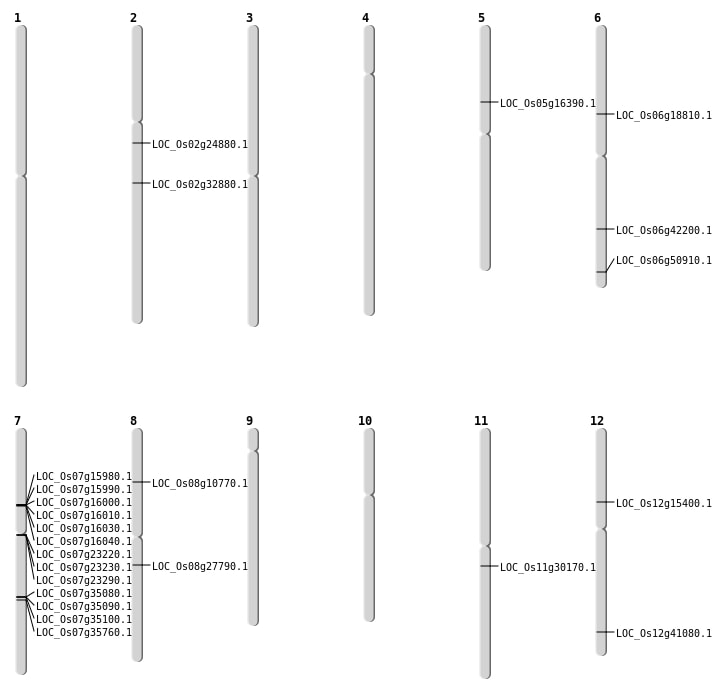
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30654149 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 20951101 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 9339915 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 11252415 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 16142767 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 17542950 | C-A | HOMO | STOP_GAINED | LOC_Os11g30170 |
| SBS | Chr11 | 9284013 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 15910129 | G-C | HOMO | INTRON | |
| SBS | Chr12 | 22091005 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 25437876 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g41080 |
| SBS | Chr2 | 15973056 | A-G | HOMO | INTRON | |
| SBS | Chr2 | 16104784 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 27868540 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 23301318 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6703434 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 9487972 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 14050171 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 24823824 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 29886601 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 8911579 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 10680127 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g18810 |
| SBS | Chr6 | 25338047 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g42200 |
| SBS | Chr6 | 28112971 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 29771975 | T-C | HET | INTRON | |
| SBS | Chr6 | 30814290 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g50910 |
| SBS | Chr6 | 30907354 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr7 | 13092163 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 13123592 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g23290 |
| SBS | Chr7 | 14123900 | G-A | HET | INTRON | |
| SBS | Chr7 | 26804921 | T-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 10171056 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 1315703 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 13636524 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 16927927 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g27790 |
| SBS | Chr9 | 15312757 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 4091276 | C-T | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 548233 | 548237 | 4 | |
| Deletion | Chr6 | 792567 | 792568 | 1 | |
| Deletion | Chr11 | 2258251 | 2258252 | 1 | |
| Deletion | Chr8 | 3346529 | 3346532 | 3 | |
| Deletion | Chr12 | 3702058 | 3702059 | 1 | |
| Deletion | Chr8 | 6332584 | 6332586 | 2 | LOC_Os08g10770 |
| Deletion | Chr9 | 6710705 | 6710710 | 5 | |
| Deletion | Chr12 | 8797140 | 8797142 | 2 | LOC_Os12g15400 |
| Deletion | Chr12 | 9017320 | 9017338 | 18 | |
| Deletion | Chr5 | 9231882 | 9231883 | 1 | |
| Deletion | Chr5 | 9279376 | 9279387 | 11 | LOC_Os05g16390 |
| Deletion | Chr7 | 9308924 | 9357532 | 48608 | 6 |
| Deletion | Chr10 | 9699861 | 9699865 | 4 | |
| Deletion | Chr7 | 13091737 | 13098058 | 6321 | 2 |
| Deletion | Chr7 | 13097240 | 13097249 | 9 | |
| Deletion | Chr2 | 14426795 | 14426803 | 8 | LOC_Os02g24880 |
| Deletion | Chr7 | 15875161 | 15875170 | 9 | |
| Deletion | Chr8 | 17761388 | 17761397 | 9 | |
| Deletion | Chr8 | 17763385 | 17763389 | 4 | |
| Deletion | Chr2 | 19533017 | 19533026 | 9 | LOC_Os02g32880 |
| Deletion | Chr7 | 21014561 | 21027795 | 13234 | 3 |
| Deletion | Chr10 | 21414178 | 21415812 | 1634 | |
| Deletion | Chr7 | 21420925 | 21424012 | 3087 | LOC_Os07g35760 |
| Deletion | Chr2 | 23745222 | 23745227 | 5 | |
| Deletion | Chr7 | 26804925 | 26804933 | 8 |
Insertions: 7
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 13098052 | 13130448 | |
| Inversion | Chr7 | 13128289 | 13130455 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 12202200 | Chr1 | 34372952 | |
| Translocation | Chr5 | 20012320 | Chr4 | 22380840 | |
| Translocation | Chr10 | 21025387 | Chr9 | 13866584 | |
| Translocation | Chr10 | 21026763 | Chr9 | 13866541 |
