Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1394-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1394-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 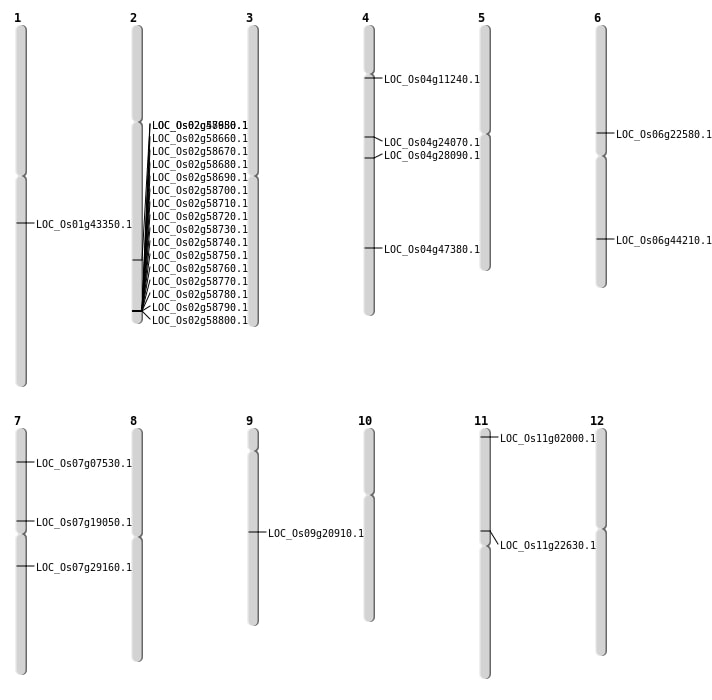
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13447419 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 17737015 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 20791210 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 3432736 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 8526520 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19242552 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 7715842 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 13013098 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g22630 |
| SBS | Chr11 | 3439128 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 634206 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 13982286 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 5792791 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 5807696 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 9334460 | G-A | HET | INTRON | |
| SBS | Chr2 | 19266649 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 27735755 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 15942948 | T-A | HOMO | INTRON | |
| SBS | Chr3 | 23287567 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 24362890 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28013903 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 30231722 | C-A | HET | INTRON | |
| SBS | Chr3 | 5365719 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 8065852 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 13749138 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24070 |
| SBS | Chr4 | 17631122 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 33496923 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 5530741 | C-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 12472657 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 2039383 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr7 | 13394288 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 17070887 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g29160 |
| SBS | Chr7 | 18213917 | C-A | HET | INTRON | |
| SBS | Chr8 | 23407762 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 25512125 | G-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 1254066 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 14135575 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 21387400 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 6775321 | C-A | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 4173253 | 4173260 | 7 | |
| Deletion | Chr3 | 5655023 | 5655024 | 1 | |
| Deletion | Chr4 | 6147462 | 6147465 | 3 | LOC_Os04g11240 |
| Deletion | Chr6 | 7447679 | 7447680 | 1 | |
| Deletion | Chr3 | 11137441 | 11137442 | 1 | |
| Deletion | Chr1 | 11261820 | 11261821 | 1 | |
| Deletion | Chr3 | 11487828 | 11487830 | 2 | |
| Deletion | Chr1 | 11658227 | 11658228 | 1 | |
| Deletion | Chr4 | 11876158 | 11876167 | 9 | |
| Deletion | Chr9 | 12583905 | 12583907 | 2 | LOC_Os09g20910 |
| Deletion | Chr6 | 13135928 | 13135931 | 3 | LOC_Os06g22580 |
| Deletion | Chr9 | 13662619 | 13662629 | 10 | |
| Deletion | Chr6 | 14094565 | 14094566 | 1 | |
| Deletion | Chr2 | 15936398 | 15936399 | 1 | |
| Deletion | Chr3 | 18103775 | 18103784 | 9 | |
| Deletion | Chr5 | 19645376 | 19645378 | 2 | |
| Deletion | Chr7 | 22301295 | 22301297 | 2 | |
| Deletion | Chr1 | 24719242 | 24719257 | 15 | |
| Deletion | Chr1 | 24784441 | 24784455 | 14 | LOC_Os01g43350 |
| Deletion | Chr11 | 25098143 | 25098144 | 1 | |
| Deletion | Chr6 | 26665343 | 26665349 | 6 | LOC_Os06g44210 |
| Deletion | Chr4 | 28117988 | 28117989 | 1 | LOC_Os04g47380 |
| Deletion | Chr2 | 28274308 | 28274313 | 5 | |
| Deletion | Chr2 | 35844001 | 35938000 | 94000 | 16 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 535206 | 535206 | 1 | LOC_Os11g02000 |
| Insertion | Chr2 | 29365387 | 29365389 | 3 | LOC_Os02g47980 |
| Insertion | Chr6 | 16147453 | 16147453 | 1 | |
| Insertion | Chr6 | 8005884 | 8005884 | 1 | |
| Insertion | Chr7 | 12674433 | 12674434 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 11273955 | 11278091 | LOC_Os07g19050 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2724268 | Chr1 | 13710389 | |
| Translocation | Chr7 | 3757718 | Chr2 | 35842954 | 2 |
| Translocation | Chr7 | 3759184 | Chr2 | 35843926 | LOC_Os02g58650 |
| Translocation | Chr4 | 16584178 | Chr2 | 35843965 | 2 |
| Translocation | Chr4 | 16584214 | Chr2 | 35842968 | 2 |
