Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1401-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1401-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 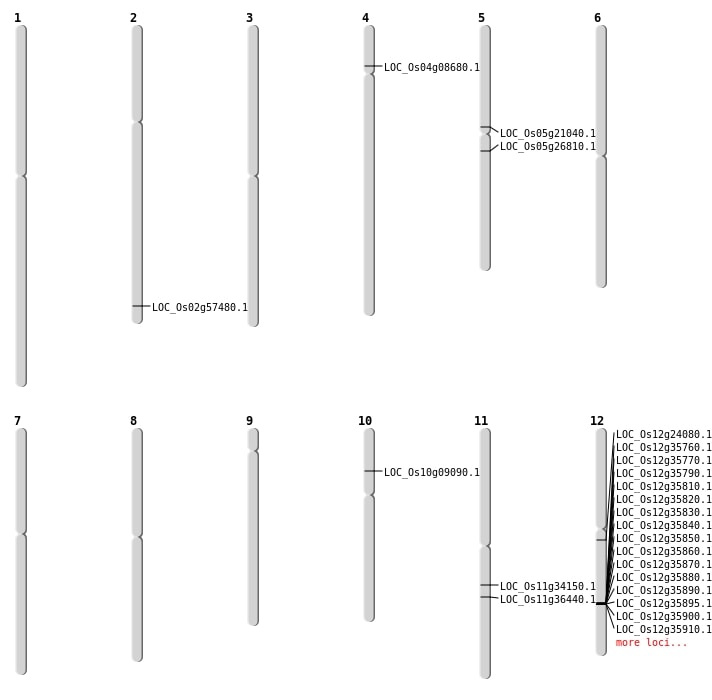
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15793169 | T-G | HET | INTRON | |
| SBS | Chr1 | 20422844 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 29542577 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 42387465 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 7764751 | C-T | HET | INTRON | |
| SBS | Chr1 | 8838973 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 11609624 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15622797 | C-T | HET | INTRON | |
| SBS | Chr10 | 7911218 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 12887052 | C-A | HET | INTRON | |
| SBS | Chr11 | 14194469 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 11842498 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 14534124 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 18112263 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr12 | 20832527 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 24357293 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 3463855 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 7406267 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 23358219 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30694258 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 6579241 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 15987267 | G-A | HET | INTRON | |
| SBS | Chr3 | 19322814 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 28552763 | C-A | HET | INTRON | |
| SBS | Chr4 | 34182348 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 4712661 | G-A | HOMO | STOP_GAINED | LOC_Os04g08680 |
| SBS | Chr5 | 13737455 | T-C | HET | INTRON | |
| SBS | Chr5 | 27224063 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 5380373 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 11732228 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 14116398 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 6200765 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 18427223 | C-T | HET | INTRON | |
| SBS | Chr8 | 20498493 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 7482310 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 11826843 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 16373493 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 22236491 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 4107808 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 7129711 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9725512 | C-G | HET | SYNONYMOUS_CODING |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 47293 | 47300 | 7 | |
| Deletion | Chr10 | 1511488 | 1511493 | 5 | |
| Deletion | Chr3 | 3681757 | 3681766 | 9 | |
| Deletion | Chr10 | 4914780 | 4914785 | 5 | LOC_Os10g09090 |
| Deletion | Chr7 | 7599067 | 7599068 | 1 | |
| Deletion | Chr9 | 9652531 | 9652546 | 15 | |
| Deletion | Chr8 | 9704316 | 9704328 | 12 | |
| Deletion | Chr11 | 10952837 | 10952838 | 1 | |
| Deletion | Chr4 | 12227093 | 12227096 | 3 | |
| Deletion | Chr11 | 15696221 | 15696223 | 2 | |
| Deletion | Chr3 | 16305754 | 16305760 | 6 | |
| Deletion | Chr7 | 18737022 | 18737029 | 7 | |
| Deletion | Chr11 | 19991888 | 19994170 | 2282 | LOC_Os11g34150 |
| Deletion | Chr9 | 21316466 | 21316467 | 1 | |
| Deletion | Chr11 | 21489746 | 21489761 | 15 | LOC_Os11g36440 |
| Deletion | Chr2 | 21684090 | 21686066 | 1976 | |
| Deletion | Chr12 | 21772001 | 21942000 | 170000 | 16 |
| Deletion | Chr2 | 21884866 | 21884869 | 3 | |
| Deletion | Chr10 | 22210412 | 22210436 | 24 | |
| Deletion | Chr2 | 23073992 | 23073993 | 1 | |
| Deletion | Chr1 | 24932604 | 24932605 | 1 | |
| Deletion | Chr7 | 25928215 | 25928219 | 4 | |
| Deletion | Chr1 | 26270394 | 26270395 | 1 | |
| Deletion | Chr3 | 26424242 | 26424243 | 1 | |
| Deletion | Chr1 | 26835621 | 26835637 | 16 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr4 | 29876670 | 29876683 | 13 | |
| Deletion | Chr2 | 35222154 | 35222162 | 8 | LOC_Os02g57480 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 12374919 | 15553099 | 2 |
| Inversion | Chr12 | 12859523 | 13699569 | LOC_Os12g24080 |
No Translocation
