Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1402-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1402-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 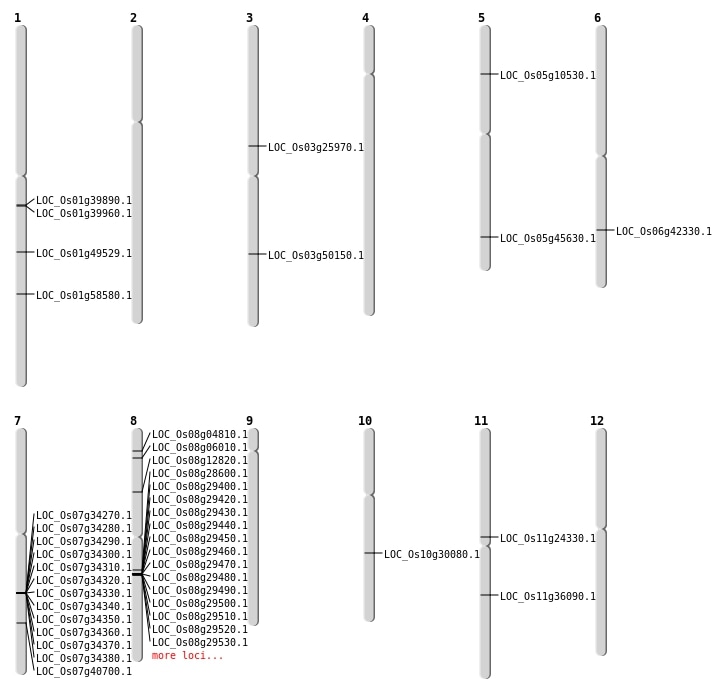
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22496312 | C-T | HET | INTRON | |
| SBS | Chr1 | 24954639 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 27863038 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 9903164 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17147353 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 18941317 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr11 | 13860190 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g24330 |
| SBS | Chr11 | 13933289 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 14196339 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 17312135 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 21215932 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g36090 |
| SBS | Chr12 | 10225568 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 1898014 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 18786646 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 16340082 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 18824819 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 22975092 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 24989988 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 24991176 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 24991177 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 32364484 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 3938180 | T-C | HET | INTRON | |
| SBS | Chr3 | 8105683 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 982835 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26894590 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 8469938 | T-G | HOMO | INTRON | |
| SBS | Chr5 | 17063136 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20981201 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 5726055 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g10530 |
| SBS | Chr6 | 10603725 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 12812975 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 23511530 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 25266108 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 12965102 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 13850780 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12662237 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 1664773 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 18028522 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g29400 |
| SBS | Chr8 | 18028523 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g29400 |
| SBS | Chr9 | 16481139 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2945844 | G-A | HET | INTRON | |
| SBS | Chr9 | 2945846 | T-A | HET | INTRON |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 2394836 | 2394841 | 5 | |
| Deletion | Chr5 | 5207922 | 5207924 | 2 | |
| Deletion | Chr8 | 7594029 | 7594030 | 1 | LOC_Os08g12820 |
| Deletion | Chr11 | 9868919 | 9868941 | 22 | |
| Deletion | Chr12 | 12964045 | 12964057 | 12 | |
| Deletion | Chr10 | 13155438 | 13155440 | 2 | |
| Deletion | Chr11 | 13473262 | 13473280 | 18 | |
| Deletion | Chr3 | 14873255 | 14873259 | 4 | LOC_Os03g25970 |
| Deletion | Chr10 | 17944359 | 17944360 | 1 | |
| Deletion | Chr8 | 18029408 | 18617031 | 587623 | 81 |
| Deletion | Chr10 | 18051150 | 18051151 | 1 | |
| Deletion | Chr8 | 18968182 | 18968214 | 32 | |
| Deletion | Chr7 | 20516001 | 20610000 | 94000 | 12 |
| Deletion | Chr12 | 20728406 | 20728407 | 1 | |
| Deletion | Chr6 | 21890069 | 21890079 | 10 | |
| Deletion | Chr3 | 22170678 | 22170680 | 2 | |
| Deletion | Chr10 | 22443393 | 22443394 | 1 | |
| Deletion | Chr7 | 24393369 | 24393370 | 1 | LOC_Os07g40700 |
| Deletion | Chr3 | 24989477 | 24989502 | 25 | |
| Deletion | Chr1 | 25167399 | 25167401 | 2 | |
| Deletion | Chr6 | 25423022 | 25423030 | 8 | LOC_Os06g42330 |
| Deletion | Chr5 | 26447216 | 26447218 | 2 | LOC_Os05g45630 |
| Deletion | Chr1 | 28484416 | 28484446 | 30 | LOC_Os01g49529 |
| Deletion | Chr3 | 28601836 | 28601857 | 21 | LOC_Os03g50150 |
| Deletion | Chr6 | 29130188 | 29130190 | 2 | |
| Deletion | Chr1 | 33870874 | 33870887 | 13 | LOC_Os01g58580 |
| Deletion | Chr3 | 36203679 | 36203681 | 2 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23218418 | 23218418 | 1 | |
| Insertion | Chr1 | 36411822 | 36411822 | 1 | |
| Insertion | Chr10 | 15621383 | 15621384 | 2 | LOC_Os10g30080 |
| Insertion | Chr11 | 2547058 | 2547058 | 1 | |
| Insertion | Chr11 | 4807214 | 4807215 | 2 | |
| Insertion | Chr4 | 5642535 | 5642535 | 1 | |
| Insertion | Chr5 | 21974748 | 21974749 | 2 | |
| Insertion | Chr6 | 29432069 | 29432070 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 2417257 | 3283954 | 2 |
| Inversion | Chr8 | 3283962 | 27671534 | 2 |
| Inversion | Chr8 | 17475762 | 18617008 | LOC_Os08g28600 |
| Inversion | Chr1 | 22498523 | 22538730 | 2 |
| Inversion | Chr1 | 22498590 | 22538757 | LOC_Os01g39890 |
No Translocation
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr10 | 17884836 | 17885061 | 225 |
