Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1403-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1403-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 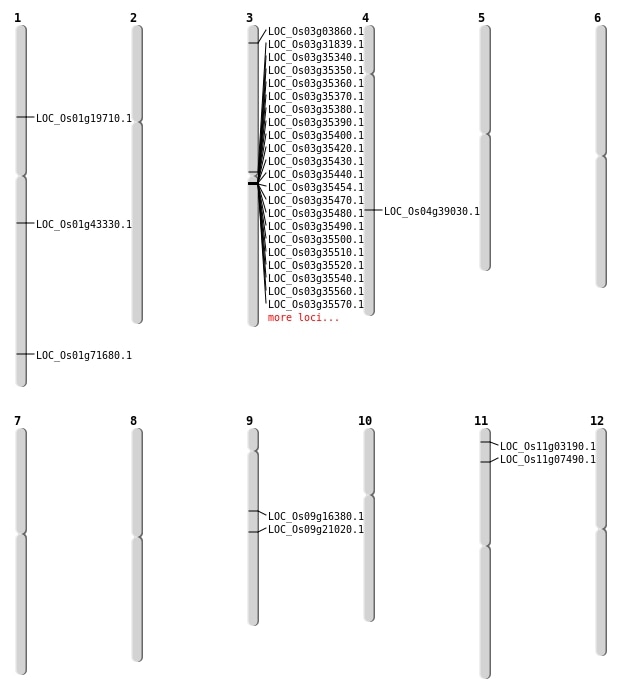
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10696927 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 39153452 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 9181635 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 17738758 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 1895170 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5139146 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 3799714 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g07490 |
| SBS | Chr12 | 12664634 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 16557131 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15515145 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 23478050 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 27685356 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 35503209 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 18279856 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 4794471 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 7128914 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 24180715 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 15316028 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15573198 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 20182443 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 28169912 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 11356737 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 7516616 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 10016690 | T-A | HOMO | STOP_GAINED | LOC_Os09g16380 |
| SBS | Chr9 | 10016693 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g16380 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 4518083 | 4518089 | 6 | |
| Deletion | Chr9 | 10020011 | 10020012 | 1 | |
| Deletion | Chr1 | 11179345 | 11179347 | 2 | LOC_Os01g19710 |
| Deletion | Chr12 | 13523557 | 13523759 | 202 | |
| Deletion | Chr9 | 13993440 | 13993447 | 7 | |
| Deletion | Chr9 | 15172626 | 15172627 | 1 | |
| Deletion | Chr3 | 15517278 | 15517281 | 3 | |
| Deletion | Chr4 | 17167515 | 17167516 | 1 | |
| Deletion | Chr10 | 17624640 | 17624656 | 16 | |
| Deletion | Chr7 | 19355115 | 19355117 | 2 | |
| Deletion | Chr3 | 19589001 | 19724000 | 135000 | 21 |
| Deletion | Chr9 | 19714487 | 19714503 | 16 | |
| Deletion | Chr3 | 19883829 | 19883837 | 8 | |
| Deletion | Chr3 | 22512455 | 22512466 | 11 | LOC_Os03g40490 |
| Deletion | Chr1 | 24766342 | 24766372 | 30 | LOC_Os01g43330 |
| Deletion | Chr12 | 26407945 | 26407946 | 1 | |
| Deletion | Chr3 | 29834923 | 29834952 | 29 | |
| Deletion | Chr1 | 30317345 | 30317360 | 15 | |
| Deletion | Chr3 | 35502209 | 35502217 | 8 | |
| Deletion | Chr1 | 38863974 | 38863975 | 1 | |
| Deletion | Chr1 | 39431442 | 39431448 | 6 | |
| Deletion | Chr1 | 41505973 | 41506002 | 29 |
Insertions: 7
Inversions: 13
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 397095 | 1160279 | LOC_Os11g03190 |
| Inversion | Chr3 | 1738688 | 2008867 | LOC_Os03g03860 |
| Inversion | Chr3 | 1738843 | 2008873 | LOC_Os03g03860 |
| Inversion | Chr9 | 10282518 | 10344417 | |
| Inversion | Chr9 | 10289200 | 12677849 | LOC_Os09g21020 |
| Inversion | Chr9 | 10344376 | 10345472 | |
| Inversion | Chr9 | 10345469 | 12680825 | LOC_Os09g21020 |
| Inversion | Chr12 | 18896803 | 26598245 | |
| Inversion | Chr3 | 21961283 | 21978264 | LOC_Os03g39580 |
| Inversion | Chr3 | 22910087 | 23467061 | |
| Inversion | Chr3 | 22910202 | 23467071 | |
| Inversion | Chr1 | 41537673 | 41990063 | LOC_Os01g71680 |
| Inversion | Chr1 | 41539794 | 41990158 | LOC_Os01g71690 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 1738706 | Chr1 | 41539793 | 2 |
| Translocation | Chr3 | 1738884 | Chr1 | 41990158 | LOC_Os03g03860 |
| Translocation | Chr10 | 2003607 | Chr4 | 23180228 | LOC_Os04g39030 |
| Translocation | Chr9 | 10282507 | Chr2 | 14996296 | |
| Translocation | Chr9 | 10287065 | Chr2 | 14996284 | |
| Translocation | Chr12 | 13523798 | Chr1 | 41539831 | LOC_Os01g71690 |
| Translocation | Chr3 | 18216853 | Chr1 | 9105546 | LOC_Os03g31839 |
